Abstract
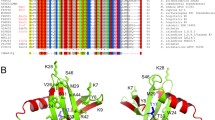
The DNA/RNA-binding KIN protein was discovered in 1989, and since then, it has been found to participate in several processes, e.g., as a transcription factor in bacteria, yeasts, and plants, in immunoglobulin isotype switching, and in the repair and resolution of double-strand breaks caused by ionizing radiation. However, the complete three-dimensional structure and biophysical properties of KIN remain important information for clarifying its function and to help elucidate mechanisms associated with it not yet completely understood. The present study provides data on phylogenetic analyses of the different domains, as well as a biophysical characterization of the human KIN protein (HSAKIN) using bioinformatics techniques, circular dichroism spectroscopy, and differential scanning calorimetry to estimate the composition of secondary structure elements; further studies were performed to determine the biophysical parameters ΔHm and Tm. The phylogenetic analysis indicated that the zinc-finger and winged helix domains are highly conserved in KIN, with mean identity of 90.37% and 65.36%, respectively. The KOW motif was conserved only among the higher eukaryotes, indicating that this motif emerged later on the evolutionary timescale. HSAKIN has more than 50% of its secondary structure composed by random coil and β-turns. The highest values of ΔHm and Tm were found at pH 7.4 suggesting a stable structure at physiological conditions. The characteristics found for HSAKIN are primarily due to its relatively low composition of α-helices and β-strands, making up less than half of the protein structure.






Similar content being viewed by others
References
Angulo JF, Moreau PL, Maunoury R, Laporte J, Hill AM, Bertolotti R, Devoret R (1989) KIN, a mammalian nuclear protein immunologically related to E. coli RecA protein. Mutat Res 217:123–134
Angulo JF, Rouer E, Benarous R, Devoret R (1991) Identification of a mouse cDNA fragment whose expressed polypeptide reacts with anti-recA antibodies. Biochimie 73:251–256
Araneda S, Angulo J, Touret M, Sallanon-Moulin M, Souchier C, Jouvet M (1997) Preferential expression of kin, a nuclear protein binding to curved DNA, in the neurons of the adult rat. Brain Res 762:103–113
Biard DS, Saintigny Y, Maratrat M, Paris F, Martin M, Angulo JF (1997) Enhanced expression of the Kin17 protein immediately after low doses of ionizing radiation. Radiat Res 147:442–450
Biard DS, Miccoli L, Despras E, Frobert Y, Creminon C, Angulo JF (2002) Ionizing radiation triggers chromatin-bound kin17 complex formation in human cells. J Biol Chem 277:19156–19165
Biard DS, Miccoli L, Despras E, Harper F, Pichard E, Creminon C, Angulo JF (2003) Participation of kin17 protein in replication factories and in other DNA transactions mediated by high molecular weight nuclear complexes. Mol Cancer Res 1:519–531
Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72:291–336
Cantor CR, Schimmel PR (1980) Biophysical chemistry. W. H Freeman, San Francisco
Carlier L, le Maire A, Braud S, Masson C, Gondry M, Zinn-Justin S, Guilhaudis L, Milazzo I, Davoust D, Gilquin B, Couprie J (2006) NMR assignment of region 51–160 of human KIN17, a DNA and RNA-binding protein. J Biomol NMR 36(Suppl 1):29
Carlier L, Couprie J, le Maire A, Guilhaudis L, Milazzo-Segalas I, Courcon M, Moutiez M, Gondry M, Davoust D, Gilquin B, Zinn-Justin S (2007) Solution structure of the region 51–160 of human KIN17 reveals an atypical winged helix domain. Protein Sci 16:2750–2755
Ceroni A, Passerini A, Vullo A, Frasconi P (2006) DISULFIND: a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res 34:W177–181
Coordinators NR (2018) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46:D8–D13
Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA (2006) Structural basis for bacterial transcription-coupled DNA repair. Cell 124:507–520
Del Sal G, Manfioletti G, Schneider C (1989) The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques 7:514–520
Despras E, Miccoli L, Creminon C, Rouillard D, Angulo JF, Biard DS (2003) Depletion of KIN17, a human DNA replication protein, increases the radiosensitivity of RKO cells. Radiat Res 159:748–758
Dunker AK, Oldfield CJ (2015) Back to the future: nuclear magnetic resonance and bioinformatics studies on intrinsically disordered proteins. Adv Exp Med Biol 870:1–34
Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN (2005) Flexible nets. The roles of intrinsic disorder in protein interaction networks. Febs J 272:5129–5148
Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Garcia-Molina A, Xing S, Huijser P (2014a) The Arabidopsis KIN17 and its homolog KLP mediate different aspects of plant growth and development. Plant Signal Behav 9:e28634
Garcia-Molina A, Xing S, Huijser P (2014b) A conserved KIN17 curved DNA-binding domain protein assembles with SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE7 to adapt Arabidopsis growth and development to limiting copper availability. Plant Physiol 164:828–840
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM (2006) Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol 2:e100
Johnson WC (1999) Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins 35:307–312
Jones S, Thornton JM (2004) Searching for functional sites in protein structures. Curr Opin Chem Biol 8:3–7
Kannouche P, Pinon-Lataillade G, Mauffrey P, Faucher C, Biard DS, Angulo JF (1997) Overexpression of kin17 protein forms intranuclear foci in mammalian cells. Biochimie 79:599–606
Kannouche P, Mauffrey P, Pinon-Lataillade G, Mattei MG, Sarasin A, Daya-Grosjean L, Angulo JF (2000) Molecular cloning and characterization of the human KIN17 cDNA encoding a component of the UVC response that is conserved among metazoans. Carcinogenesis 21:1701–1710
Kay BK (2012) SH3 domains come of age. FEBS Lett 586:2606–2608
Kyrpides NC, Woese CR, Ouzounis CA (1996) KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci 21:425–426
Le MX, Haddad D, Ling AK, Li C, So CC, Chopra A, Hu R, Angulo JF, Moffat J, Martin A (2016) Kin17 facilitates multiple double-strand break repair pathways that govern B cell class switching. Sci Rep 6:37215
le Maire A, Schiltz M, Braud S, Gondry M, Charbonnier JB, Zinn-Justin S, Stura E (2006) Crystallization and halide phasing of the C-terminal domain of human KIN17. Acta Crystallogr Sect F Struct Biol Cryst Commun 62:245–248
le Maire A, Schiltz M, Stura EA, Pinon-Lataillade G, Couprie J, Moutiez M, Gondry M, Angulo JF, Zinn-Justin S (2006) A tandem of SH3-like domains participates in RNA binding in KIN17, a human protein activated in response to genotoxics. J Mol Biol 364:764–776
Masson C, Menaa F, Pinon-Lataillade G, Frobert Y, Chevillard S, Radicella JP, Sarasin A, Angulo JF (2003) Global genome repair is required to activate KIN17, a UVC-responsive gene involved in DNA replication. Proc Natl Acad Sci USA 100:616–621
Mazin A, Milot E, Devoret R, Chartrand P (1994a) KIN17, a mouse nuclear protein, binds to bent DNA fragments that are found at illegitimate recombination junctions in mammalian cells. Mol Gen Genet MGG 244:435–438
Mazin A, Timchenko T, Menissier-de Murcia J, Schreiber V, Angulo JF, de Murcia G, Devoret R (1994b) Kin17, a mouse nuclear zinc finger protein that binds preferentially to curved DNA. Nucleic Acids Res 22:4335–4341
McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R (2013) Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41:W597–600
Miccoli L, Biard DS, Creminon C, Angulo JF (2002) Human kin17 protein directly interacts with the simian virus 40 large T antigen and inhibits DNA replication. Cancer Res 62:5425–5435
Miccoli L, Frouin I, Novac O, Di Paola D, Harper F, Zannis-Hadjopoulos M, Maga G, Biard DS, Angulo JF (2005) The human stress-activated protein kin17 belongs to the multiprotein DNA replication complex and associates in vivo with mammalian replication origins. Mol Cell Biol 25:3814–3830
Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK (2008) Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genom 9(Suppl 1):S1
Olsson U, Wolf-Watz M (2010) Overlap between folding and functional energy landscapes for adenylate kinase conformational change. Nat Commun 1:111
Paiardini A, Bossa F, Pascarella S (2004) Evolutionarily conserved regions and hydrophobic contacts at the superfamily level: the case of the fold-type I, pyridoxal-5ʹ-phosphate-dependent enzymes. Protein Sci 13:2992–3005
Patil A, Nakamura H (2006) Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett 580:2041–2045
Pils B, Copley RR, Schultz J (2005) Variation in structural location and amino acid conservation of functional sites in protein domain families. BMC Bioinform 6:210
Pinon-Lataillade G, Masson C, Bernardino-Sgherri J, Henriot V, Mauffrey P, Frobert Y, Araneda S, Angulo JF (2004) KIN17 encodes an RNA-binding protein and is expressed during mouse spermatogenesis. J Cell Sci 117:3691–3702
Ramos AC, Gaspar VP, Kelmer SM, Sellani TA, Batista AG, De Lima Neto QA, Rodrigues EG, Fernandez MA (2015) The kin17 Protein in Murine Melanoma Cells. Int J Mol Sci 16:27912–27920
Riener CK, Kada G, Gruber HJ (2002) Quick measurement of protein sulfhydryls with Ellman's reagent and with 4,4ʹ-dithiodipyridine. Anal Bioanal Chem 373:266–276
Saksela K, Permi P (2012) SH3 domain ligand binding: What's the consensus and where's the specificity? FEBS Lett 586:2609–2614
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Singh R, Valcarcel J (2005) Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol 12:645–653
Sreerama N, Woody RW (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem 287:252–260
Steiner T, Kaiser JT, Marinkovic S, Huber R, Wahl MC (2002) Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. Embo J 21:4641–4653
Timchenko T, Bailone A, Devoret R (1996) Btcd, a mouse protein that binds to curved DNA, can substitute in Escherichia coli for H-NS, a bacterial nucleoid protein. Embo J 15:3986–3992
Tompa P (2005) The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett 579:3346–3354
UniProt Consortium T (2018) UniProt: the universal protein knowledgebase. Nucleic Acids Res 46:2699
Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA (2004) Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol 14:208–216
Vucetic S, Xie H, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN (2007) Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res 6:1899–1916
Wedemeyer WJ, Welker E, Narayan M, Scheraga HA (2000) Disulfide bonds and protein folding. Biochemistry 39:4207–4216
Westermeier R, Naven T (2002) Part II: course manual, step 9: ingel digestion. In: Proteomics in practice: a laboratory manual of proteome analysis. Wiley-VCH, Weinheim, p 261
Wetzel R, Becker M, Behlke J, Billwitz H, Bohm S, Ebert B, Hamann H, Krumbiegel J, Lassmann G (1980) Temperature behaviour of human serum albumin. Eur J Biochem 104:469–478
Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112:531–552
Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN (2007a) Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res 6:1917–1932
Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z (2007b) Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res 6:1882–1898
Yang YL, Suen J, Brynildsen MP, Galbraith SJ, Liao JC (2005) Inferring yeast cell cycle regulators and interactions using transcription factor activities. BMC Genom 6:90
Zhang Y, Gao H, Gao X, Huang S, Wu K, Yu X, Yuan K, Zeng T (2017a) Elevated expression of Kin17 in cervical cancer and its association with cancer cell proliferation and invasion. Int J Gynecol Cancer 27:628–633
Zhang Y, Huang S, Gao H, Wu K, Ouyang X, Zhu Z, Yu X, Zeng T (2017b) Upregulation of KIN17 is associated with non-small cell lung cancer invasiveness. Oncol Lett 13:2274–2280
Acknowledgements
This work was supported by Fundação Araucária (Grant Numbers 147/14 and 40/16), Coordination for the Improvement of Higher Education Personnel—Brazil (CAPES, code 001), and the National Council for Scientific and Technological Development—Brazil (CNPq Grant Number 305960/2015-6). Dr. Fátima Pereira de Souza and Dr. Marcelo Andrés Fossey are gratefully acknowledged for the helpful discussion, and the authors thank IBILCE/UNESP and UFPR for the use of facilities.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pattaro Júnior, J.R., Caruso, Í.P., de Lima Neto, Q.A. et al. Biophysical characterization and molecular phylogeny of human KIN protein. Eur Biophys J 48, 645–657 (2019). https://doi.org/10.1007/s00249-019-01390-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-019-01390-3