Abstract
Drug-resistant Mycobacterium tuberculosis (M. tuberculosis) has become an increasingly serious public health problem and has complicated tuberculosis (TB) treatment. Levofloxacin (LOF) is an ideal anti-tuberculosis drug in clinical applications. However, the detailed molecular mechanisms of LOF-resistant M. tuberculosis in TB treatment have not been revealed. Our study performed transcriptome and methylome sequencing to investigate the potential biological characteristics of LOF resistance in M. tuberculosis H37Rv. In the transcriptome analysis, 953 differentially expressed genes (DEGs) were identified; 514 and 439 DEGs were significantly downregulated and upregulated in the LOF-resistant group and control group, respectively. The KEGG pathway analysis revealed that 97 pathways were enriched in this study. In the methylome analysis, 239 differentially methylated genes (DMGs) were identified; 150 and 89 DMGs were hypomethylated and hypermethylated in the LOF-resistant group and control group, respectively. The KEGG pathway analysis revealed that 74 pathways were enriched in this study. The overlap study suggested that 25 genes were obtained. It was notable that nine genes expressed downregulated mRNA and upregulated methylated levels, including pgi, fadE4, php, cyp132, pckA, rpmB1, pfkB, acg, and ctpF, especially cyp132, pckA, and pfkB, which were vital in LOF-resistant M. tuberculosis H37Rv. The overlapping genes between transcriptome and methylome could be essential for studying the molecular mechanisms of LOF-resistant M. tuberculosis H37Rv. These results may provide informative evidence for TB treatment with LOF.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is an infectious disease usually caused by Mycobacterium tuberculosis (M. tuberculosis) infection (Zaman 2010). According to a World Health Organization (WHO) report, there were 10 million new cases of TB and 1.3 million deaths as a result of TB in 2017. In India, China, and Indonesia, the total number of people with TB accounts for 44% of the cases worldwide. In China, about 889,000 TB cases were reported, accounting for 9% of the cases worldwide (WHO 2018). Therefore, the prevention of TB remains an urgent task. In recent years, the global TB epidemic has declined. Multidrug-resistant TB (MDR-TB) refers to TB that is resistant to two or more anti-TB drugs, such as rifampicin and isoniazid (Seung et al. 2015). The clinical abuse of anti-TB drugs has led to an increasing number of drug-resistant strains. Moreover, the failure rate of MDR-TB treatment is significantly higher than that at which susceptible bacteria are infected with TB (Ormerod 2005). As such, the development and testing of new drugs are direly needed to enable more timely and effective treatment of MDR-TB.
Levofloxacin (LOF) is a third-generation fluorinated quinolone antibiotic that is an optically active L-isomer of the racemate ofloxacin (Moorthy et al. 2008). LOF plays an important role in inhibiting the activity of the M. tuberculosis gyrase, which severely blocks the replication process of DNA. Therefore, this drug could effectively promote DNA degradation and kill bacteria in a short period time (Collin et al. 2011). The bactericidal activity of this molecule against intracellular and extracellular M. tuberculosis could be more than twice that of ofloxacin (McGregor et al. 2008). In recent years, it has been clinically proposed that LOF can directly inhibit the metabolism of Mycolic acid in mycobacteria (Ghimire et al. 2016). Thus, it plays a therapeutic role in treating TB. In clinical practice, LOF is an ideal anti-TB drug without any cross-resistance in combination with other clinically applied anti-TB drugs (Richeldi et al. 2002). However, evidence of the detailed molecular mechanisms of LOF resistance in TB is still insufficient.
The main process of DNA methylation is the conversion of the 5′-terminal cytosine of CpG dinucleotide to a 5ʹ-methylcytosine due to the activity of DNA methyltransferase and a methylated CpG-binding protein (Lister et al. 2009). There is evidence that DNA methylation modifications are closely associated with a variety of diseases (Robertson 2005), including tumorigenesis, infectious diseases, autoimmune diseases, chronic diseases, and blood diseases (Ramos et al. 2011; Saleem et al. 2015; Zhuang et al. 2012). Moreover, previous studies have highlighted occurrence of DNA methylation remodeling in dendritic cells upon infection with M. tuberculosis (Pacis et al. 2019, 2015). In addition, researchers have focused on the relationship between drug resistance and the M. tuberculosis complex (MTC) (Madsen et al. 2005; Phunpruch et al. 2013). For example, Phunpruch et al. suggested that the methylation pattern in Mycobacterium smegmatis (M. smegmatis) could be used to understand the resistance toward lincosamides and erythromycin (Phunpruch et al. 2013). Therefore, we speculate that DNA methylation may be closely related to LOF resistance in the M. tuberculosis H37Rv strain.
In this study, we have employed LOF to study the drug resistance in the M. tuberculosis H37Rv strain. The transcriptomes and methylomes of the LOF-resistant M. tuberculosis H37Rv strain and the wild-type M. tuberculosis H37Rv strain were studied using a high-throughput sequencing method. Differentially expressed genes (DEGs) and methylated genes (DMGs) have been investigated. KEGG pathway analysis has revealed the important signal pathways related to LOF resistance. In addition, a molecular regulation network has been constructed based on the DEGs and DMGs. The results provide useful information regarding LOF resistance in M. tuberculosis H37Rv.
Materials and Methods
Sample Treatment
In this study, the wild-type M. tuberculosis H37Rv strain was obtained from the Sample Bank of the Reference Laboratory of Guangdong Province and preserved in Löwenstein–Jensen (LJ) medium (Thermo Fisher Scientific, USA) at 37 °C for 4 weeks. Subsequently, a monoclone was chosen. The monoclonal agglomerate was dispersed and diluted to 1 Mech turbidity using a bacterial ultrasonic dispersion counter. The prepared bacterial solution was inoculated with LJ medium for amplification. This generation of strain was designated as the primary G0. The WHO criteria (Guidelines for surveillance of drug resistance in tuberculosis. WHO Geneva/IUATLD Paris. International Union Against Tuberculosis and Lung Disease 1998) for LOF-resistant strains suggested that M. tuberculosis H37Rv strains could survive in a medium containing LOF at 2.0 μg/mL. Therefore, the G0 strain was further cultured in LJ medium at 37 °C for 4 weeks with 2.0 μg/mL LOF (Sigma-Aldrich, USA). The drug-resistant strain was named the G1 generation. The same steps were repeated until G4 strains obtained, which satisfied the criteria provided by the WHO. Subsequently, three more generations of the drug-resistant G4 strains and the wild-type M. tuberculosis H37Rv strain were cultured under identical conditions (LJ medium at 37 °C for 4 weeks per generation). LOF-resistant G7 strains and the wild-type M. tuberculosis H37Rv strain were designated as the LOF-resistant group and control group, respectively. Bacteria in both groups were stored at − 80 °C for further DNA and RNA extraction.
Transcriptome
The total RNAs of bacteria in the LOF-resistant and control groups were extracted using a MiniBEST Universal RNA Extraction Kit (Takara, Japan). Purity and concentration were determined using NanoDrop ND1000 (FisherScientific, USA). DNase I (Epicenter, USA) was used to remove DNA contamination. Ribosomal RNA was removed using an Epicenter Ribo-zero™ rRNA Removal Kit (Epicenter, USA). Linear RNA was fragmented into 180 bp using the ultrasonic method (Covaris M220, USA). After fragmentation and random priming, reverse transcription was carried out using a PrimeScript™ II Strand cDNA Synthesis Kit (Takara, Japan). An NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB,USA) was used according to the manufacturer’s protocol. After the end repair, blunting, and adenylation of 3ʹ ends of cDNA fragments, an adaptor was ligated to each cDNA end. Subsequently, size selection (approximately 400 bp) was carried out using an AMPure XP system (Beckman, USA). Then, adaptor-ligated cDNA was amplified with universal PCR primers. The final library products were purified and assessed using an Agilent Bioanalyzer 2100 system (Agilent, USA). The clustering of the index-coded sample was performed using a cBot Cluster Generation System (Illumina, USA). The libraries were sequenced on an Illumina HiSeq 2500 platform with the 150-bp paired-end sequencing strategy, and the raw data were finally obtained. Subsequently, high-quality reads were obtained after the removal of low-quality reads (sequences containing the linker, > 10% N bases, or > 50% bases with a < 10 mass value). The aligned reads were obtained by mapping high-quality reads in the M. tuberculosis H37Rv genome (https://www.ncbi.nlm.nih.gov/genome/166?genome_assembly_id=159857) using HISAT (version 0.1.6, -phred33 -p 3 -x * -known-splicesite-infile *). The aligned reads were further mapped onto the gene exon region via HTseq (version 0.6.1, -count -f bam -r pos -s no -i gene_name). Finally, the mapped reads were used for differential expression analysis via DESeq (version 2) combined with R (version 3.1) under default parameters (Love et al. 2014).
Methylome
DNA of 1 × 106 bacteria cells in the LOF-resistant and control groups were obtained using a QIAamp DNA Mini Kit (Qiagen, USA). 100U MspI (NEB, USA) was used to digest the 5 μg genomic DNA from the two groups for 16 h at 37 °C. An Illumina sequencing library was constructed using a paired-end sequencing strategy. In brief, library preparation includes three main steps: end repair, “A” base addition, and methylated-adapter ligation. The size distribution of the library was examined with 2% agarose gel. A MinElute PCR Purification Kit (QIAGEN, USA) was used to recover DNA by columns using a 20 µl elution buffer. Bisulfite conversion was performed using an EZ DNA Methylation-Gold Kit (ZYMO, USA). Bisulfite-treated products were amplified by a 50 µl PCR reaction system, which contained 10 µl-treated DNA, 1 µl 10 mM dNTP, 1 µl PCR primer A, 1 µl PCR primer B, 5 µl 10 × master mix buffer, 0.5 µl JumpStartTM Taq DNA polymerase, and 31.5 µl water. The PCR program was implemented as follows: 94 °C 1 min, 11 cycles (94 °C 0.5 min, 58 °C 0.5 min, and 72 °C 0.5 min), 72 °C 5 min. The PCR products were purified and recovered with a QIAamp DNA Mini Kit (Qiagen, USA) followed by sequencing with Illumina HiSeq2500 with the 150-bp paired-end sequencing strategy. BGI SOAPaligner version 2.01 with two mismatches for successful mapping was used to identify the strand specificity of DNA methylation (Li et al. 2009). CpG loci with significant differences between the two groups in DNA methylation have been identified through surrogate variable analysis (Leek and Storey 2007).
Bioinformatics
A heat map of DEGs and DMGs was created using the Cluster 3.0 software (USA) (FDR ≤ 0.001 and |log2 ratio| ≥ 1). KEGG signaling pathway analysis was conducted using the KOBAS software (version 3.0, https://kobas.cbi.pku.edu.cn/). The protein–protein interaction network analysis was based on the Search Tool for the Retrieval of Interacting Genes (STRING) (version 11.0, https://string-db.org/).
Statistics
Data were expressed as mean ± SD. Spearman’s linear regression analysis was applied to identify the correlation between differential methylation and gene expression levels. A probability value of P < 0.05 was considered statistically significant.
Results
Transcriptome
In this study, the results suggested that 953 genes could be retrieved to be differentially expressed between the LOF-resistant and control groups (Fig. 1a and Table S1). Of these 953 DEGs, in the LOF-resistant group, 514 were significantly downregulated, and 439 were significantly upregulated. Moreover, all 953 DEGs were involved in 97 KEGG pathways and enriched in this study. In Fig. 1b and Table S2, enriched metabolic pathways (mtu01100) with 111 DEGs, biosynthesis of secondary metabolites (mtu01110) with 49 DEGs, and microbial metabolism in diverse environments (mtu01120) with 43 DEGs were presented; these pathways are significant to the further study of LOF resistance in M. tuberculosis H37Rv using transcriptome analysis.
Transcriptome atlas and KEGG pathway analysis of DEGs between the LOF-resistant group and control group. a Heatmap of DEGs between the LOF-resistant group and control group. b KEGG pathway analysis of DEGs between the LOF-resistant group and control group. DEGs differentially expressed genes, LOF levofloxacin
Methylome
In this study, 239 DMGs were identified (Table S3). In the LOF-resistant group, 150 DMGs were hypomethylated, and 89 DMGs were hypermethylated (Fig. 2a). Furthermore, the KEGG pathway analysis of these DMGs was conducted. The results indicate that all 239 DMGs were involved in 74 KEGG pathways (Fig. 2b and Table S4), including metabolic pathways (mtu01100) with 67 DMGs, biosynthesis of secondary metabolites (mtu01110) with 32 DMGs, and biosynthesis of antibiotics (mtu01130) with 25 DMGs; these pathways are significant to the further study of LOF resistance in M. tuberculosis H37Rv under methylome analysis.
Methylome atlas and KEGG pathway analysis of DMGs between the LOF-resistant group and control group. a Heatmap of DMGs between the LOF-resistant group and control group. b KEGG pathway analysis of DMGs between the LOF-resistant group and control group. DMGs differentially methylated genes, LOF levofloxacin
Overlap of Transcriptome and Methylome and Function Analysis
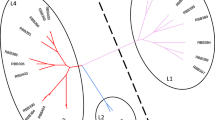
In order to obtain the overlapping genes of the DEGs and DMGs, we compared the two datasets. We obtained 25 overlapping genes (Table S5) after analyzing these two datasets. Of these 25 genes, 16 genes expressed upregulation and hypomethylation in the LOF-resistant group compared with those in the control group. Nine genes expressed downregulation and hypermethylation in the LOF-resistant group compared with those in the control group. These results were consistent with a previous study indicating that DNA hypomethylated and hypermethylated genes could promote and inhibit gene expression (Moore et al. 2013). In this study, we examined the associations among 25 overlapping genes between transcriptome and methylome. The results indicate that DMGs were negatively correlated with mRNA abundance (Fig. 3), which quantitatively verifies the negative relationship between methylation and gene expression in the LOF-resistant M. tuberculosis H37Rv strain.
In order to further study the functions of DMGs, all 25 genes were searched against the STRING database, as presented in Figure S1. It is notable that the hypermethylated genes fadE4 (Rv0231), php (Rv0230c), pgi (Rv0946c), pckA (Rv0211), and pfkB (Rv2029c) interacted within various protein–protein interaction modules (Fig. 4). Additionally, these 25 genes were subjected to the KEGG pathway analysis, from which 25 signaling pathways were obtained (Table S6), including biosynthesis of antibiotics (mtu01130) with five genes and biosynthesis of secondary metabolites (mtu01110) with five genes; these pathways are significant to the further study of LOF resistance in M. tuberculosis H37Rv using a combination of transcriptome and methylome analysis. The result of protein–protein interaction (PPI) analysis would provide potential information regarding the associations of various M. tuberculosis proteins with levofloxacin resistance, which are valuable and warrant further research.
Analysis of protein–protein interaction networks of nine hypermethylated genes. The protein interaction networks for nine differentially methylated genes between the LOF-resistant group and control group. LOF levofloxacin; red, hypermethylated genes; green, hypomethylated genes; blue, other interacting proteins
Discussion
The treatment of MDR-TB is a difficult problem in current TB control. MDR-TB treatment has obvious disadvantages, such as high cost, low cure rate, and high mortality rate. Therefore, MDR-TB poses a significant threat to public health (Migliori et al. 2010). LOF has been proven effective in treating TB, which has been the subject of clinical trials to improve dosage amounts (Tiberi et al. 2018). According to the results obtained by Ghimire et al., there are several advantages of using LOF to treat MDR-TB. First, the concentration of LOF in tissues, such as sputum, bronchial mucosa, and lung tissue, could exceed that in the blood. Second, LOF could easily kill M. tuberculosis in macrophages. Third, continuous delivery of this drug has no accumulation tendency (Ghimire et al. 2016). Therefore, LOF could be an optimal drug for MDR-TB treatments and thus is in phase II trials (NTC01918397). However, the basic molecular mechanisms of LOF resistance in TB treatment have not been fully demonstrated.
In this study, we have studied transcriptome and methylome atlases of LOF-resistant M. tuberculosis H37Rv compared with the control group. The results suggested that 953 DEGs and 239 DMGs were obtained between the LOF-resistant and control groups. This result suggests that LOF resistance affects the change of internal regulation networks in M. tuberculosis H37Rv. Epigenetic abnormalities provide an alternative mechanism for transcriptional silencing. Therefore, we have focused on genes with downregulated mRNA expressions and upregulated methylation levels in both the LOF-resistant and control groups. Nine genes were identified: pgi (Rv0946c), fadE4 (Rv0231), php (Rv0230c), cyp132 (Rv1394c), pckA (Rv0211), rpmB1 (Rv0105c), pfkB (Rv2029c), acg (Rv2032), and ctpF (Rv1997).
In M. tuberculosis H37Rv, pgi (Rv0946c) encodes a protein belonging to glucose-6-phosphate isomerase, which catalyzes the interconversion of d-glucopyranose 6-phosphate and β-d-fructofuranose 6-phosphate. This process is an essential step of the glycolysis and gluconeogenesis pathways. Apart from its enzymatic functions, pgi (Rv0946c) also has a number of other functions, including being an autocrine motility factor (AMF), a neuroleukin (NLK) agent, a serine proteinase inhibitor, and a differentiation and maturation mediator. As in most organisms, pgi (Rv0946c) is essential for the survival of M. tuberculosis in macrophages (Sassetti et al. 2003). Furthermore, a previous study demonstrated that cyp132 (Rv1394c) plays a crucial role in Rv1395’s regulator function in cytochrome P450 regulation in M. tuberculosis (Recchi et al. 2003). Recchi et al. (2003) suggested that Rv1395 could bind to two adjacent sites located between two divergent genes: a cytochrome P450 gene—cyp132 (Rv1394c) and the Rv1395 gene itself. Rv1395 could induce the expression of the cytochrome P450 gene (cyp132) and repress its own transcription. Previous studies of metabolism in M. tuberculosis suggested that pckA (Rv0211) in M. tuberculosis can be related to anaplerotic and gluconeogenic routes. This suggests that pckA (Rv0211) might be related to the regulation of metabolic adaptations of central metabolism in various environments of M. tuberculosis (Beste et al. 2011; Watanabe et al. 2011). Moreover, as a member of the DOS regulator, the expression of pfkB (Rv2029c) is upregulated in hypoxia conditions and macrophages (Shi et al. 2016). The results of the mutation screening revealed that pfkB (Rv2029c) is not related to M. tuberculosis growth in vitro or in vivo (Phong et al. 2013; Sassetti and Rubin 2003). A previous study revealed that this gene encodes 6-phosphate fructokinase (pfkB), which is involved in the conversion of sugar 1-p into sugar 1,6-P (Phong et al. 2013). Recently, this protein was identified as a potential vaccine candidate (Zvi et al. 2008). Therefore, pfkB (Rv2029c) is potentially essential in vaccine development in M. tuberculosis treatment. Combined with the network of protein–protein interaction, we can also confirm that the hypermethylated genes cyp132 (Rv1394c), pckA (Rv0211), and pfkB (Rv2029c) were essential in M. tuberculosis H37Rv with LOF resistance. The above genes were both enriched in biosynthesis of antibiotics (mtu01130) and biosynthesis of secondary metabolites (mtu01110) pathways in the LOF-resistant M. tuberculosis H37Rv group, which could be essential for the future study of M. tuberculosis treatment. In summary, multiple molecular mechanisms could facilitate in understanding the LOF-resistant mechanisms of M. tuberculosis H37Rv when mediated by methylation. Each of the genes identified in this study could be essential for future treatments of MDR-TB.
In this study, 953 genes were differentially expressed (LOF-resistant group/control group), metabolic pathways (mtu01100) was the most enriched in the KEGG pathway analysis. A total of 239 genes were differentially methylated (LOF-resistant group/control group), metabolic pathways (mtu01100) was also the most enriched in the KEGG pathway analysis. A total of 25 genes were overlapping between the transcriptome and methylome datasets, which may be the potential core genes of LOF function in M. tuberculosis. There were nine genes with downregulated mRNA expression and hypermethylation levels in the LOF-resistant group: pgi, fadE4, php, cyp132, pckA, rpmB1, pfkB, acg, and ctpF; in particular, cyp132, pckA, and pfkB deserve more in-depth study. The overlapping genes between transcriptome and methylome could be essential for studying the molecular mechanisms of LOF-resistant M. tuberculosis H37Rv. The above results could provide informative evidence for TB treatment using LOF.
Data Availability
Raw sequencing data presented in this paper are available under the GEO accession number PRJNA579441.
References
Beste DJV et al (2011) 13C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog 7:e1002091
Collin F, Karkare S, Maxwell A (2011) Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Appl Microbiol Biotechnol 92:479–497. https://doi.org/10.1007/s00253-011-3557-z
Ghimire S et al (2016) Pharmacokinetic/pharmacodynamic-based optimization of levofloxacin administration in the treatment of MDR-TB. J Antimicrob Chemother 71:2691–2703. https://doi.org/10.1093/jac/dkw164
Leek JT, Storey JD (2007) Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 3:1724–1735. https://doi.org/10.1371/journal.pgen.0030161
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics (Oxford, England) 25:1966–1967. https://doi.org/10.1093/bioinformatics/btp336
Lister R et al (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322. https://doi.org/10.1038/nature08514
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Madsen CT, Jakobsen L, Douthwaite S (2005) Mycobacterium smegmatis Erm(38) is a reluctant dimethyltransferase. Antimicrob Agents Chemother 49:3803–3809. https://doi.org/10.1128/aac.49.9.3803-3809.2005
McGregor JC, Allen GP, Bearden DT (2008) Levofloxacin in the treatment of complicated urinary tract infections and acute pyelonephritis. Therap Clin Risk Manag 4:843–853. https://doi.org/10.2147/tcrm.s3426
Migliori GB, Centis R, Lange C, Richardson MD, Sotgiu G (2010) Emerging epidemic of drug-resistant tuberculosis in Europe, Russia, China, South America and Asia: current status and global perspectives. Curr Opin Pulmonary Med 16:171–179. https://doi.org/10.1097/MCP.0b013e328337573e
Moore LD, Le T, Fan G (2013) DNA methylation and its basic function. Neuropsychopharmacology 38:23–38. https://doi.org/10.1038/npp.2012.112
Moorthy N, Raghavendra N, Venkatarathnamma PN (2008) Levofloxacin-induced acute psychosis. Indian J Psychiatry 50:57–58. https://doi.org/10.4103/0019-5545.39762
Ormerod LP (2005) Multidrug-resistant tuberculosis (MDR-TB): epidemiology, prevention and treatment. Br Med Bull 73–74:17–24. https://doi.org/10.1093/bmb/ldh047
Pacis A et al (2019) Gene activation precedes DNA demethylation in response to infection in human dendritic cells. Proc Natl Acad Sci 116:6938. https://doi.org/10.1073/pnas.1814700116
Pacis A et al (2015) Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. https://doi.org/10.1101/gr.192005.115
Phong WY, Lin W, Rao SP, Dick T, Alonso S, Pethe K (2013) Characterization of phosphofructokinase activity in Mycobacterium tuberculosis reveals that a functional glycolytic carbon flow is necessary to limit the accumulation of toxic metabolic intermediates under hypoxia. PLoS ONE 8:e56037. https://doi.org/10.1371/journal.pone.0056037
Phunpruch S, Warit S, Suksamran R, Billamas P, Jaitrong S, Palittapongarnpim P, Prammananan T (2013) A role for 16S rRNA dimethyltransferase (ksgA) in intrinsic clarithromycin resistance in Mycobacterium tuberculosis. Int J Antimicrob Agents 41:548–551. https://doi.org/10.1016/j.ijantimicag.2013.02.011
Ramos EA et al (2011) Epigenetic changes of CXCR4 and its ligand CXCL12 as prognostic factors for sporadic breast cancer. PLoS ONE 6:e29461. https://doi.org/10.1371/journal.pone.0029461
Recchi C, Sclavi B, Rauzier J, Gicquel B, Reyrat JM (2003) Mycobacterium tuberculosis Rv1395 is a class III transcriptional regulator of the AraC family involved in cytochrome P450 regulation. J Biol Chem 278:33763–33773. https://doi.org/10.1074/jbc.M305963200
Richeldi L et al (2002) Clinical use of Levofloxacin in the long-term treatment of drug resistant tuberculosis. Monaldi Arch Chest Dis 57:39–43
Robertson KD (2005) DNA methylation and human disease. Nat Rev Genetics 6:597–610. https://doi.org/10.1038/nrg1655
Saleem M et al (2015) Review-epigenetic therapy for cancer. Pak J Pharm Sci 28:1023–1032
Sassetti CM, Boyd DH, Rubin EJ (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84
Sassetti CM, Rubin EJ (2003) Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA 100:12989–12994. https://doi.org/10.1073/pnas.2134250100
Seung KJ, Keshavjee S, Rich ML (2015) Multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis. Cold Spring Harb Perspect Med 5:a017863. https://doi.org/10.1101/cshperspect.a017863
Shi L et al (2016) Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Mol Microbiol 99:1179. https://doi.org/10.1111/mmi.13350
Tiberi S, Munoz-Torrico M, Duarte R, Dalcolmo M, D'Ambrosio L, Migliori GB (2018) New drugs and perspectives for new anti-tuberculosis regimens. Pulmonology 24:86–98. https://doi.org/10.1016/j.rppnen.2017.10.009
Watanabe S, Zimmermann M, Goodwin MB, Sauer U, Barry CE 3rd, Boshoff HI (2011) Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog 7:e1002287. https://doi.org/10.1371/journal.ppat.1002287
WHO (2018) Global tuberculosis report 2018. WHO, Geneva
WHO Geneva/IUATLD Paris, International Union Against Tuberculosis and Lung Disease (1998) Guidelines for surveillance of drug resistance in tuberculosis. Int J Tuberc Lung Dis 2:72–89
Zaman K (2010) Tuberculosis: a global health problem. J Health Popul Nutr 28:111–113
Zhuang J et al (2012) The dynamics and prognostic potential of DNA methylation changes at stem cell gene loci in women's cancer. PLoS Genet 8:e1002517. https://doi.org/10.1371/journal.pgen.1002517
Zvi A, Ariel N, Fulkerson J, Sadoff JC, Shafferman A (2008) Whole genome identification of Mycobacterium tuberculosis vaccine candidates by comprehensive data mining and bioinformatic analyses. BMC Med Genom 1:18. https://doi.org/10.1186/1755-8794-1-18
Acknowledgements
Authors would like to thank Science and Technology Planning Project of Guangdong Province, China (2016A020216021), Science and Technology Planning Project of Guangzhou (201604020006), and National Science and Technology Key Projects on “Major Infectious Diseases such as HIV/AIDS, Viral Hepatitis Prevention and Treatment” (2018ZX10715004-002, 2017ZX10201302-005, 2018ZX10103001-001–004) for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
All authors declare no conflict of interest.
Additional information
Handling editor: Ashley Teufel.
Hai-cheng Li and Hui-xin Guo contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
239_2019_9926_MOESM1_ESM.tif
Figure S1. The networks of protein–protein interactions for 25 overlapping differentially methylated genes between the LOF-resistant group and control group. LOF levofloxacin; red, hypermethylated genes; green, hypomethylated genes; blue, other interacted proteins. (TIF 2979 kb)
239_2019_9926_MOESM2_ESM.xls
Table S1. Differentially expressed genes between the LOF-resistant group and control group. LOF levofloxacin. (XLS 178 kb)
239_2019_9926_MOESM3_ESM.xls
Table S2. KEGG pathway analysis of differentially expressed genes between the LOF-resistant group and control group. LOF levofloxacin. (XLS 67 kb)
239_2019_9926_MOESM4_ESM.xls
Table S3. Differentially methylated genes between the LOF-resistant group and control group. LOF levofloxacin. (XLS 41 kb)
239_2019_9926_MOESM5_ESM.xls
Table S4. KEGG pathway analysis of differentially methylated genes between the LOF-resistant group and control group. LOF levofloxacin. (XLS 52 kb)
239_2019_9926_MOESM6_ESM.xls
Table S5. Overlapping genes of differentially expressed genes and differentially methylated genes between the LOF-resistant group and control group. LOF levofloxacin. (XLS 24 kb)
239_2019_9926_MOESM7_ESM.xls
Table S6. KEGG pathway analysis of overlapping genes between the LOF-resistant group and control group. LOF levofloxacin. (XLS 34 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Hc., Guo, Hx., Chen, T. et al. Potential Genes Related to Levofloxacin Resistance in Mycobacterium tuberculosis Based on Transcriptome and Methylome Overlap Analysis. J Mol Evol 88, 202–209 (2020). https://doi.org/10.1007/s00239-019-09926-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-019-09926-z