Abstract
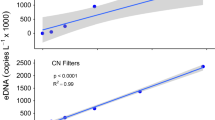
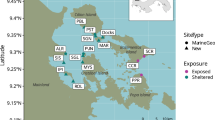
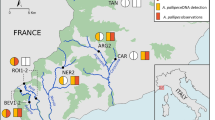
Environmental DNA (eDNA) is increasingly being used in aquatic environments for monitoring species, particularly those that are of conservation concern and/or are difficult to visually observe. Quantitative PCR (qPCR) has been employed to detect low abundance species occurring in environmental water samples. However, the qPCR technique has principally been applied to freshwater habitats, with less application to pelagic marine environments. We developed a species-specific eDNA assay for the Chilean devil ray, Mobula tarapacana, to assess the capability of using eDNA to detect transient pelagic marine animals. For this pilot study, seawater samples taken at seamounts around the Azores (NE Atlantic) were tested to determine the suitability of this approach for detecting the target species. Samples were taken at sites where M. tarapacana has been previously observed, in addition to sites where its presence is not known. eDNA detection was compared to observations carried out on the same day as water sampling. The qPCR assay successfully detected M. tarapacana at four of five seamount sampling opportunities where the species was observed, and there is a statistically significant relationship between genetic and visual detection. Target DNA was found at one location between seamounts in the absence of visual observation. Our results highlight the importance of physical environmental factors in relation to sampling eDNA in the ocean, such as currents and eDNA dispersal ability. This method has been shown to be sensitive for detection of M. tarapacana DNA in seawater and, therefore, in the identification of important seamounts requiring conservation.




Similar content being viewed by others
References
Bourlat SJ, Borja A, Gilbert J, Taylor MI, Davies N, Weisberg SB, Griffith JF, Lettieri T, Field D, Benzie J, Glöckner FO, Rodríguez-Ezpeleta N, Faith DP, Bean TP, Obst M (2013) Genomics in marine monitoring: new opportunities forassessing marine health status. Mar Pollut Bull 74:19–31
Bylemans J, Furlan EM, Pearce L, Daly T, Gleeson DM (2016) Improving the containment of a freshwater invader using environmental DNA (eDNA) based monitoring. Biol Invasions 18:3081–3089
Couturier LIE, Marshall AD, Jaine FRA, Kashiwagi T, Pierce SJ, Townsend KA, Weeks SJ, Bennett MB, Richardson AJ (2012) Biology, ecology and conservation of the Mobulidae. J Fish Biol 80:1075–1119
Davison PI, Creach V, Liang W-J, Andreou D, Britton JR, Copp GH (2016) Laboratory and field validation of a simple method for detecting four species of non-native freshwater fish using eDNA. J Fish Biol 89:1782–1793
Deiner K, Altermatt F (2014) Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 9:e88786
Dejean T, Valentini A, Duparc A, Pellier-Cuit S, Pompanon F, Taberlet P, Miaud C (2011) Persistence of environmental DNA in freshwater ecosystems. PLoS ONE 6(8):e23398
Dudgeon CL, Blower DC, Broderick D, Giles JL, Holmes BJ, Kashiwagi T, Krück NC, Morgan JAT, Tillett BJ, Ovendon JR (2012) A review of the application of molecular genetics for fisheries management and conservation of sharks and rays. J Fish Biol 80:1789–1843
Ficetola GF, Miaud C, Pompanon F, Taberlet P (2008) Species detection using environmental DNA from water samples. Biol Lett 4:423–425
Foote AD, Thomsen PF, Sveegaard S, Wahlberg M, Kielgast J, Kyhn LA, Salling AB, Galatius A, Orlando L, Gilbert MTP (2012) Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS ONE 7(8):e41781
Goldberg CS, Turner CR, Deiner K, Klymus KE, Thomsen PF, Murphy MA, Spear SF, McKee A, Oyler-McCance SJ, Cornman RJ, Laramie MB, Mahon AR, Lance RF, Pilliod DS, Strickler KM, Waits LP, Fremier AK, Takahara T, Herder JE, Taberlet P (2016) Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol Evol 7:1299–1307
Gustavson MS, Collins PC, Finarelli JA, Egan D, Conchúir R, Wightman GD, King JJ, Gauthier DT, Whelan K, Carlsson JEL, Carlsson J (2015) An eDNA assay for Irish Petromyzon marinus and Salmo trutta and field validation in running water. J Fish Biol 87:1254–1262
Herder JE, Valentini A, Bellemain E, Dejean T, van Delft JJCW, Thomsen PF, Taberlet P (2014) Environmental DNA – a review of the possible applications for the detection of (invasive) species. Stichting RAVON, Nijmegen. Report, pp 2013–2104
Hoelzel A (2001) Shark fishing in fin soup. Conserv Genet 2:69–72
Holmes BH, Steinke D, Ward RD (2009) Identification of shark and ray fins using DNA barcoding. Fish Res 95:280–288
Jerde CL, Mahon AR, Chadderton WL, Lodge DM (2011) “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv Lett 4:150–157
Kelly RP, Port JA, Yamahara KM, Crowder LB (2014) Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE 9:e86175
Kutyavin IV, Afonina IA, Lukhtanov Mills A, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, Dempcy R, Reed MW, Meyer RB, Meyer J (2000) 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res 28:655–661
Laramie MB, Pilliod DS, Goldberg CS (2014) Characterizing the distribution of an endangered salmonid using environmental DNA analysis. Biol Conserv 183:29–37
Morato T, Varkey DA, Damaso C et al (2008a) Evidence of a seamount effect on aggregating visitors. Mar Ecol Prog Ser 357:23–32
Morato T, Machete M, Kitchingman A et al (2008b) Abundance and distribution of seamounts in the Azores. Mar Ecol Prog Ser 357:17–21
Morato T, Hoyle SD, Allain V, Nicol SJ (2010) Seamounts are hotspots of pelagic biodiversity in the open ocean. Proc Natl Acad Sci USA 107:9707–9711
Pardo SA, Walls RHL, Bigman JS (2016) Mobula tarapacana. (errata version published in 2016) The IUCN Red List of Threatened Species 2016: e.T60199A100016302. Accessed 08 Jan 2017
Piaggio AJ, Engeman RM, Hopken MW, Humphrey JS, Keacher KL, Bruce WE, Avery ML (2014) Detecting an elusive invasive species: a diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Mol Ecol Res 14:374–380
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2014) Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol Ecol Res 14:109–116
Pitcher TJ, Morato T, Hart PJB, Clark MR, Haggan N, Santos RS (2007) The depths of ignorance an ecosystem evaluation framework for seamount ecology, fisheries, and conservation. In: Pitcher TJ, Morato T, Hart PJB, Clark MR, Haggan N, Santos RS (eds) Seamounts: ecology, fisheries, and conservation. Blackwell Fisheries and Aquatic Resources Series 12. Blackwell Publishing, Oxford, pp 476–488
Poortvliet M, Olsen JL, Croll DA, Bernardi G, Newton K, Kollias S, O’Sullivan J, Fernando D, Stevens G, Galván Magaña F, Seret B, Wintner S, Hoarau G (2015) A dated molecular phylogeny of manta and devil rays (Mobulidae) based on mitogenome and nuclear sequences. Mol Phyl Evol 83:72–85
Shaw JLA, Clarke LJ, Wedderburn SD et al (2016) Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biol Conserv 197:131–138
Sigsgaard EE, Nielsen IB, Bach SS, Lorenzen ED, Robinson DP, Knudsen SW, Pedersen MW, Jaidah MA, Orlando L, Willerslev E, Møller PR, Thomsen PF (2016) Population characteristics of a large whale shark aggregation inferred from seawater environmental DNA. Nat Ecol Evol 1:1–4
Simpfendorfer CA, Kyne PM, Noble TH, Goldsbury J, Basiita RK, Lindsay R, Shields A, Perry C, Jerry DR (2016) Environmental DNA detects Critically Endangered largetooth sawfish in the wild. Endanger Species Res 30:109–116
Sobral AF, Afonso P (2014) Occurrence of mobulids in the Azores, central North Atlantic. J Mar Biol Assoc United Kingdom 94(08):1671–1675
Strickler KM, Fremier AK, Goldberg CS (2015) Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol Conserv 183:85–92
Taberlet P, Coissac E, Hajibabaei M, Rieseberg LH (2012) Environmental DNA. Mol Ecol 21:1789–1793
Takahara T, Minamoto T (2012) Doi H Estimation of fish biomass using environmental DNA. PLoS ONE 8:e56584
Thomsen PF, Kielgast J, Iversen LL, Møller PR, Rasmussen M, Willerslev E (2012) Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 7:e41732
Thorrold SR, Afonso P, Fontes J, Braun CD, Santos RS, Skomal GB et al (2014) Extreme diving behaviour in devil rays links surface waters and the deep ocean. Nat Commun. doi:10.1038/ncomms5274
Tinti F, Ungaro N, Pasolini P et al (2003) Development of molecular and morphological markers to improve species-specific monitoring and systematics of Northeast Atlantic and Mediterranean skates (Rajiformes). J Exp Mar Biol Ecol 288:149–165
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia's fish species. Philos Trans R Soc B: Biol Sci 360(1462):1847–1857
Acknowledgements
This project is part of LMG’s Ph.D., with funding from University College Dublin. Additional funding was made available by IFCT Exploratory Project (IF/01194/2013/CP1199/CT0002) and ACORES-01-0145-FEDER-00056 MAPGes. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant agreement No. 678760 (ATLAS). This output reflects only the author’s view and the European Union cannot be held responsible for any use that may be made of the information contained therein. The authors would like to thank Damian Egan for assistance in primer design and qPCR optimisation. They also gratefully acknowledge Edward Farrell and Bernard Seret for providing the tissue samples used to optimise the qPCR assay, as well as Pedro Afonso, Jorge Fontes and Sérgio Gomes for Mobula tarapacana tissue samples and for assistance with water sample collection. TM is supported by Program Investigador FCT (IF/01194/2013), FSE and MCTES, through POPH and QREN. They also thank the reviewers for their helpful comments and suggestions to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Responsible Editor: T. Reusch.
Reviewed by J. Shaw and an undisclosed expert.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gargan, L.M., Morato, T., Pham, C.K. et al. Development of a sensitive detection method to survey pelagic biodiversity using eDNA and quantitative PCR: a case study of devil ray at seamounts. Mar Biol 164, 112 (2017). https://doi.org/10.1007/s00227-017-3141-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-017-3141-x