Abstract
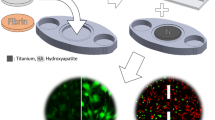
We present an insert-based approach to fabricate scalable and multiplexable microfluidic devices for 3D cell culture and integration with downstream detection modules. Laser-cut inserts with a layer of electrospun fibers are used as a scaffold for 3D cell culture, with the inserts being easily assembled in a 3D-printed fluidic device for flow-based studies. With this approach, the number and types of cells (on the inserts) in one fluidic device can be customized. Moreover, after an investigation (i.e., stimulation) under flowing conditions, the cell-laden inserts can be removed easily for subsequent studies including imaging and cell lysis. In this paper, we first discuss the fabrication of the device and characterization of the fibrous inserts. Two device designs containing two (channel width = 260 μm) and four (channel width = 180 μm) inserts, respectively, were used for different experiments in this study. Cell adhesion on the inserts with flowing media through the device was tested by culturing endothelial cells. Macrophages were cultured and stimulated under different conditions, the results of which indicate that the fibrous scaffolds under flow conditions result in dramatic effects on the amount and kinetics of TNF-α production (after LPS stimulation). Finally, we show that the cell module can be integrated with a downstream absorbance detection scheme. Overall, this technology represents a new and versatile way to culture cells in a more in vivo fashion for in vitro studies with online detection modules.

This paper describes an insert-based microfluidic device for 3D cell culture that can be easily scaled, multiplexed, and integrated with downstream analytical modules.







Similar content being viewed by others
References
Mehling M, Tay S. Microfluidic cell culture. Curr Opin Biotechnol. 2014;25:95–102.
Selimovic A, Erkal JL, Spence DM, Martin RS. Microfluidic device with tunable post arrays and integrated electrodes for studying cellular release. Analyst. 2014;139(22):5686–94.
Johnson AS, Selimovic A, Martin RS. Microchip-based electrochemical detection for monitoring cellular systems. Anal Bioanal Chem. 2013;405(10):3013–20.
Menon NV, Chuah YJ, Cao B, Lim M, Kang YJ. A microfluidic co-culture system to monitor tumor-stromal interactions on a chip. Biomicrofluidics. 2014;8(6).
Selimovic S, Dokmeci MR, Khademhosseini A. Organs-on-a-chip for drug discovery. Curr Opin Pharmacol. 2013;13(5):829–33.
Petersen NJ, Mogensen KB, Kutter JP. Performance of an in-plane detection cell with integrated waveguides for UV/Vis absorbance measurements on microfluidic separation devices. Electrophoresis. 2002;23(20):3528–36.
Sassa F, Morimoto K, Satoh W, Suzuki H. Electrochemical techniques for microfluidic applications. Electrophoresis. 2008;29(9):1787–800.
Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, Monuki ES, et al. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip. 2005;5(4):401–6.
Bennett MR, Pang WL, Ostroff NA, Baumgartner BL, Nayak S, Tsimring LS, et al. Metabolic gene regulation in a dynamically changing environment. Nature. 2008;454(7208):1119–22.
Shao JB, Wu L, Wu JZ, Zheng YH, Zhao H, Jin QH, et al. Integrated microfluidic chip for endothelial cells culture and analysis exposed to a pulsatile and oscillatory shear stress. Lab Chip. 2009;9(21):3118–25.
van der Meer AD, Poot AA, Feijen J, Vermes I. Analyzing shear stress-induced alignment of actin filaments in endothelial cells with a microfluidic assay. Biomicrofluidics. 2010;4(1)
Sung KE, Su G, Pehlke C, Trier SM, Eliceiri KW, Keely PJ, et al. Control of 3-dimensional collagen matrix polymerization for reproducible human mammary fibroblast cell culture in microfluidic devices. Biomaterials. 2009;30(27):4833–41.
Sell S, Barnes C, Smith M, McClure M, Madurantakam P, Grant J, et al. Extracellular matrix regenerated: tissue engineering via electrospun biomimetic nanofibers. Polym Int. 2007;56(11):1349–60.
Yang F, Murugan R, Ramakrishna S, Wang X, Ma YX, Wang S. Fabrication of nano-structured porous PLLA scaffold intended for nerve tissue engineering. Biomaterials. 2004;25(10):1891–900.
Naba A, Pearce OMT, Del Rosario A, Ma DD, Ding HM, Rajeeve V, et al. Characterization of the extracellular matrix of normal and diseased tissues using proteomics. J Proteome Res. 2017;16(8):3083–91.
Schlaepfer DD, Hunter T. Signal transduction from the extracellular matrix—a role for the focal adhesion protein-tyrosine kinase FAK. Cell Struct Funct. 1996;21(5):445–50.
Sheetz MP, Felsenfeld DP, Galbraith CG. Cell migration: regulation of force on extracellular-matrix-integrin complexes. Trends Cell Biol. 1998;8(2):51–4.
Mo XM, Xu CY, Kotaki M, Ramakrishna S. Electrospun P(LLA-CL) nanofiber: a biomimetic extracellular matrix for smooth muscle cell and endothelial cell proliferation. Biomaterials. 2004;25(10):1883–90.
Lebaron RG, Athanasiou KA. Extracellular matrix cell adhesion peptides: functional applications in orthopedic materials. Tissue Eng. 2000;6(2):85–103.
Zhang WJ, Zhang YS, Bakht SM, Aleman J, Shin SR, Yue K, et al. Elastomeric free-form blood vessels for interconnecting organs on chip systems. Lab Chip. 2016;16(9):1579–86.
Chen YF, Chan HN, Michael SA, Shen YS, Chen Y, Tian Q, et al. A microfluidic circulatory system integrated with capillary-assisted pressure sensors. Lab Chip. 2017;17(4):653–62.
Skardal A, Shupe T, Atala A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov Today. 2016;21(9):1399–411.
Liu YL, Chen CP, Summers S, Medawala W, Spence DM. C-peptide and zinc delivery to erythrocytes requires the presence of albumin: implications in diabetes explored with a 3D-printed fluidic device. Integr Biol. 2015;7(5):534–43.
Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21(12):745–54.
Theberge AB, Yu JQ, Young EWK, Ricke WA, Bushman W, Beebe DJ. Microfluidic multiculture assay to analyze biomolecular signaling in angiogenesis. Anal Chem. 2015;87(6):3239–46.
Golden AP, Tien J. Fabrication of microfluidic hydrogels using molded gelatin as a sacrificial element. Lab Chip. 2007;7(6):720–5.
Pham QP, Sharma U, Mikos AG. Electrospinning of polymeric nanofibers for tissue engineering applications: a review. Tissue Eng. 2006;12(5):1197–211.
Greiner A, Wendorff JH. Electrospinning: a fascinating method for the preparation of ultrathin fibres. Angew Chem Int Edit. 2007;46(30):5670–703.
Sill TJ, von Recum HA. Electrospinning: applications in drug delivery and tissue engineering. Biomaterials. 2008;29(13):1989–2006.
Li WJ, Laurencin CT, Caterson EJ, Tuan RS, Ko FK. Electrospun nanofibrous structure: a novel scaffold for tissue engineering. J Biomed Mater Res. 2002;60(4):613–21.
Chen CP, Mehl BT, Sell SA, Martin RS. Use of electrospinning and dynamic air focusing to create three-dimensional cell culture scaffolds in microfluidic devices. Analyst. 2016;141(18):5311–20.
Yang YM, Chen XM, Ding F, Zhang PY, Liu J, Go XS. Biocompatibility evaluation of silk fibroin with peripheral nerve tissues and cells in vitro. Biomaterials. 2007;28(9):1643–52.
Doe WF, Henson PM. Macrophage stimulation by bacterial lipopolysaccharides. I. Cytolytic effect on tumor target cells. J Exp Med. 1978;148(2):544–56.
Rago R, Mitchen J, Wilding G. DNA fluorometric assay in 96-well tissue-culture plates using Hoechst-33258 after cell-lysis by freezing in distilled water. Anal Biochem. 1990;191(1):31–4.
Fujiwara N, Kobayashi K. Macrophages in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4(3):281–6.
Arango Duque G, Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol. 2014;5:491.
Tabas I. Macrophage death and defective inflammation resolution in atherosclerosis. Nat Rev Immunol. 2010;10(1):36–46.
Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–96.
Chensue SW, Terebuh PD, Remick DG, Scales WE, Kunkel SL. In vivo biologic and immunohistochemical analysis of interleukin-1 alpha, beta and tumor necrosis factor during experimental endotoxemia. Kinetics, Kupffer cell expression, and glucocorticoid effects. Am J Pathol. 1991;138(2):395–402.
Vignery A. Macrophage fusion: are somatic and cancer cells possible partners? Trends Cell Biol. 2005;15(4):188–93.
Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281(5379):1001–5.
Garg K, Pullen NA, Oskeritzian CA, Ryan JJ, Bowlin GL. Macrophage functional polarization (M1/M2) in response to varying fiber and pore dimensions of electrospun scaffolds. Biomaterials. 2013;34(18):4439–51.
Au AK, Huynh W, Horowitz LF, Folch A. 3D-printed microfluidics. Angew Chem Int Edit. 2016;55(12):3862–81.
Chen CP, Mehl BT, Munshi AS, Townsend AD, Spence DM, Martin RS. 3D-printed microfluidic devices: fabrication, advantages and limitations—a mini review. Anal Meth. 2016;8(31):6005–12.
MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–50.
Lancaster JR. A tutorial on the diffusibility and reactivity of free nitric oxide. Nitric Oxide. 1997;1(1):18–30.
Ivanov VM. The 125th anniversary of the Griess reagent. J Anal Chem. 2004;59(10):1002–5.
Acknowledgements
Support from the National Institute of General Medical Sciences (Award Number R15GM084470-04) is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 1280 kb)
Rights and permissions
About this article
Cite this article
Chen, C., Townsend, A.D., Hayter, E.A. et al. Insert-based microfluidics for 3D cell culture with analysis. Anal Bioanal Chem 410, 3025–3035 (2018). https://doi.org/10.1007/s00216-018-0985-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-0985-y