Abstract
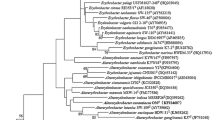
A polyphasic taxonomic study was performed on strain KMU-34T, which was isolated from seawater in the Republic of Korea. The bacterial cells were Gram-stain-negative, strictly aerobic, chemoheterotrophic, yellow-pigmented and rod-shaped. Phylogenetic analysis based on the 16S rRNA gene sequence indicated that the novel marine strain was affiliated with the family Erythrobacteraceae of the class Alphaproteobacteria and that it showed highest sequence similarity (98.6%) to Erythrobacter atlanticus s21-N3T. The DNA–DNA relatedness values between strains KMU-34T and E. atlanticus KCTC 42697T were 8.6 ± 1.2%. The DNA G + C content of strain KMU-34T was determined to be 60.4 mol%. Ubiquinone 10 (Q-10) was the sole respiratory quinone. The predominant cellular fatty acids were C18:1 ω7c (43.8%) and C16:1 ω7c (16.8%). Strain KMU-34T had phosphatidylethanolamine, phosphatidylglycerol, phosphatidylcholine, a sphingoglycolipid, an unidentified phospholipid and two unidentified lipids as polar lipids. From the distinct phylogenetic position and combination of genotypic and phenotypic characteristics, the strain is considered to represent a novel species of the genus Erythrobacter for which the name Erythrobacter alti sp. nov. is proposed. The type strain of E. alti sp. nov. is KMU-34T (= KCCM 90261T = NBRC 111903T).

Similar content being viewed by others
References
Collins MD, Jones D (1981) A note on the separation of natural mixtures of bacterial ubiquinones using reverse-phase partition thin-layer chromatography and high performance liquid chromatography. J Appl Bacteriol 51:129–134
Denner EBM, Vybiral D, Koblízek M, Kämpfer P, Busse HJ, Velimirov B (2002) Erythrobacter citreus sp. nov., a yellow-pigmented bacterium that lacks bacteriochlorophyll a, isolated from the western Mediterranean Sea. Int J Syst Evol Microbiol 52:1655–1661
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Towards defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Fuerst JA, Hawkins JA, Holmes A, Sly LI, Moore CJ, Stackebrandt E (1993) Porphyrobacter neustonensis gen. nov., sp. nov., an aerobic bacteriochlorophyll-synthesizing budding bacterium from fresh water. Int J Syst Bacteriol 43:125–134
Garrity GM, Brenner DJ, Krieg NR, Staley JT (2005) Bergey’s manual of systematic bacteriology, vol 2, 2nd edn. Springer, New York
Giovannoni SJ, Rappé M (2000) Evolution, diversity, and molecular ecology of marine procaryotes. In: Kirchman DL (ed) In microbial ecology of the oceans. Wiley, New York, pp 47–84
Hansen GH, Sørheim R (1991) Improved method for phenotypical characterization of marine bacteria. J Microbiol Methods 13:231–241
Ivanova EP, Bowman JP, Lysenko AM, Zhukova NV, Gorshkova NM, Kuznetsova TA, Kalinovskaya NI, Shevchenko LS, Mikhailov VV (2005) Erythrobacter vulgaris sp. nov., a novel organism isolated from the marine invertebrates. Syst Appl Microbiol 28:123–130
Jung YT, Park S, Oh TK, Yoon JH (2012) Erythrobacter marinus sp. nov., isolated from seawater. Int J Syst Evol Microbiol 62:2050–2055
Jung YT, Park S, Lee JS, Yoon JH (2014) Erythrobacter lutimaris sp. nov., isolated from a tidal flat sediment. Int J Syst Evol Microbiol 64:4184–4190
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through the comparative studies of sequence evolution. J Mol Evol 16:111–120
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kwon KK, Woo JH, Yang SH, Kang JH, Kang SG, Kim SJ, Sato T, Kato C et al (2007) Altererythrobacter epoxidivorans gen. nov., sp. nov., an epoxide hydrolase-active, mesophilic marine bacterium isolated from cold-seep sediment, and reclassification of Erythrobacter luteolus Yoon et al. 2005 as Altererythrobacter luteolus comb. nov. Int J Syst Evol Microbiol 57:2207–2211
Lee KB, Liu CT, Anzai Y, Kim H, Aono T, Oyaizu H (2005) The hierarchical system of the ‘Alphaproteobacteria’: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int J Syst Evol Microbiol 55:1907–1919
Lee YS, Lee DH, Kahng HY, Kim EM, Jung JS (2010) Erythrobacter gangjinensis sp. nov., a marine bacterium isolated from seawater. Int J Syst Evol Microbiol 60:1413–1417
Lei X, Zhang H, Chen Y, Li Y, Chen Z, Lai Q, Zhang J, Zheng W, Xu H, Zheng T (2015) Erythrobacter luteus sp. nov., isolated from mangrove sediment. Int J Syst Evol Microbiol 65:2472–2478
Lewin RA, Lounsbery DM (1969) Isolation, cultivation and characterization of flexibacteria. J Gen Microbiol 58:145–170
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G + C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinines and polar lipids. J Microbiol Methods 2:233–241
Power DA, Johnson JA (2009) Difco™ and BBL™ manual: manual of microbiological culture media, 2nd edn. Becton Dickinson and Company, Sparks, pp 359–360
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc, Newark
Shiba T, Simidu U (1982) Erythrobacter longus gen. nov., sp. nov., an aerobic bacterium which contains bacteriochlorophyll a. Int J Syst Evol Microbiol 32:211–217
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Stackebrandt E, Murray RGE, Trüper HG (1988) Proteobacteria classis nov., a name for the phylogenetic taxon that includes the ‘‘purple bacteria and their relatives’’. Int J Syst Bacteriol 38:321–325
Subhash Y, Tushar L, Sasikala Ch, Ramana CV (2013) Erythrobacter odishensis sp. nov. and Pontibacter odishensis sp. nov. isolated from dry soil of a solar saltern. Int J Syst Evol Microbiol 63:4524–4532
Suzuki K, Kaneko T, Komagata K (1981) Deoxyribonucleic acid homologies among coryneform bacteria. Int J Syst Bacteriol 31:131–138
Tamura K, Peterson D, Petersen N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Worliczek HL, Kämpfer P, Rosengarten R, Tindall RBJ, Busse HJ (2007) Polar lipid and fatty acid profiles-re-vitalizing old approaches as a modern tool for the classification of mycoplasmas? Syst Appl Microbiol 30:355–370
Xu XW, Wu YH, Wang CS, Wang XG, Oren A, Wu M (2009) Croceicoccus marinus gen. nov., sp. nov., a yellow-pigmented bacterium from deep-sea sediment, and emended description of the family Erythrobacteraceae. Int J Syst Evol Microbiol 59:2247–2253
Xu M, Xin Y, Yu Y, Zhang J, Zhou Y, Liu H, Tian J, Li Y (2010) Erythrobacter nanhaisediminis sp. nov., isolated from marine sediment of the South China Sea. Int J Syst Evol Microbiol 60:2215–2220
Yoon JH, Kang KH, Oh TK, Park YH (2004) Erythrobacter aquimaris sp. nov., isolated from sea water of a tidal flat of the Yellow Sea in Korea. Int J Syst Evol Microbiol 54:1981–1985
Yoon BJ, Lee DH, Oh DC (2013) Erythrobacter jejuensis sp. nov., isolated from seawater. Int J Syst Evol Microbiol 63:1421–1426
Yoon J, Lee KC, Lee JS (2016) Cribrihabitans pelagius sp. nov., a marine alphaproteobacterium isolated from seawater. Int J Syst Evol Microbiol 66:3195–3200
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. doi:10.1099/ijsem.0.001755
Yurkov V, Stackebrandt E, Holmes A, Fuerst JA, Hugenholtz P, Golecki J, Gad’on N, Gorlenko VM, Kompantseva EI, Drews G (1994) Phylogenetic positions of novel aerobic, bacteriochlorophyll a-containing bacteria and description of Roseococcus thiosulfatophilus gen. nov., sp. nov., Erythromicrobium ramosum gen. nov., sp. nov., and Erythrobacter litoralis sp. nov. Int J Syst Bacteriol 44:427–434
Acknowledgments
This research was supported by the Bisa Research Grant of Keimyung University in 2016.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
203_2017_1384_MOESM1_ESM.pptx
Supplementary Fig. 1. Transmission electron micrograph of a negatively stained cell of strain KMU-34T. Bar, 500 nm (PPTX 1192 kb)
203_2017_1384_MOESM2_ESM.pptx
Supplementary Fig. 2. Thin-layer chromatograms showing the total polar lipid compositions of KMU-34T. Total polar lipids were detected by spraying the plate with molybdatophosphoric acid, molybdenun blue, α-naphthol and ninhydrin. PE: phosphatidylethanolamine, PG: phosphatidylglycerol, PC: phosphatidylcholine, SGL: sphingoglycolipid, UPL: unidentified phospholipid, UL: unidentified lipid (PPTX 401 kb)
Rights and permissions
About this article
Cite this article
Yoon, J. Erythrobacter alti sp. nov., a marine alphaproteobacterium isolated from seawater. Arch Microbiol 199, 1133–1139 (2017). https://doi.org/10.1007/s00203-017-1384-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-017-1384-z