Abstract
Aims/hypothesis
Epigenetic regulation of gene expression has been implicated in the pathogenesis of obesity and type 2 diabetes. However, detailed information, such as key transcription factors in pancreatic beta cells that mediate environmental effects, is not yet available.
Methods
To analyse genome-wide cis-regulatory profiles and transcriptome of pancreatic islets derived from a diet-induced obesity (DIO) mouse model, we conducted chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-Seq) of histone H3 lysine 27 acetylation (histone H3K27ac) and high-throughput RNA sequencing. Transcription factor-binding motifs enriched in differential H3K27ac regions were examined by de novo motif analysis. For the predicted transcription factors, loss of function experiments were performed by transfecting specific siRNA in INS-1, a rat beta cell line, with and without palmitate treatment. Epigenomic and transcriptional changes of possible target genes were evaluated by ChIP and quantitative RT-PCR.
Results
After long-term feeding with a high-fat diet, C57BL/6J mice were obese and mildly glucose intolerant. Among 39,350 islet cis-regulatory regions, 13,369 and 4610 elements showed increase and decrease in ChIP-Seq signals, respectively, significantly associated with global change in gene expression. Remarkably, increased H3K27ac showed a distinctive genomic localisation, mainly in the proximal-promoter regions, revealing enriched elements for nuclear respiratory factor 1 (NRF1), GA repeat binding protein α (GABPA) and myocyte enhancer factor 2A (MEF2A) by de novo motif analysis, whereas decreased H3K27ac was enriched for v-maf musculoaponeurotic fibrosarcoma oncogene family protein K (MAFK), a known negative regulator of beta cells. By siRNA-mediated knockdown of NRF1, GABPA or MEF2A we found that INS-1 cells exhibited downregulation of fatty acid β-oxidation genes in parallel with decrease in the associated H3K27ac. Furthermore, in line with the epigenome in DIO mice, palmitate treatment caused increase in H3K27ac and induction of β-oxidation genes; these responses were blunted when NRF1, GABPA or MEF2A were suppressed.
Conclusions/interpretation
These results suggest novel roles for DNA-binding proteins and fatty acid signalling in obesity-induced epigenomic regulation of beta cell function.
Data availability
The next-generation sequencing data in the present study were deposited at ArrayExpress.
RNA-Seq:
Dataset name: ERR2538129 (Control), ERR2538130 (Diet-induced obesity)
Repository name and number: E-MTAB-6718 - RNA-Seq of pancreatic islets derived from mice fed a long-term high-fat diet against chow-fed controls.
ChIP-Seq:
Dataset name: ERR2538131 (Control), ERR2538132 (Diet-induced obesity)
Repository name and number: E-MTAB-6719 - H3K27ac ChIP-Seq of pancreatic islets derived from mice fed a long-term high-fat diet (HFD) against chow-fed controls.
Similar content being viewed by others

Introduction
Type 2 diabetes and obesity are chronic metabolic disorders that affect an increasing number of people globally [1, 2]. Genome-wide association studies (GWAS) [3] have highlighted the role of cis-regulatory elements in various disease processes by combining knowledge of comprehensive epigenetic profiling [4,5,6]. However, given the modest effect of identified susceptibility variants, achieving a full understanding of a disease remains challenging [3].
There is clear evidence supporting the role of environmental factors in type 2 diabetes [1, 2, 7]. Obesity, along with reduced physical activity, is regarded as being responsible for the recent pandemic [7]. Insufficient insulin secretion is essential in the development of type 2 diabetes, and a variety of environmental factors as well as genetic predisposition play important roles in the modification of beta cell function [7].
Much remains unknown, especially in humans, about the molecular characteristics of the pancreatic islets of Langerhans under various physiological and pathological conditions [8]. Several studies have provided new insights through more comprehensive analyses [4, 9,10,11]. Among assays for epigenomic profiling, histone H3 lysine 27 acetylation (H3K27ac), a marker of active regulatory regions [12], is recognised as being valuable not only in annotating human variants [6] but also in providing information on diseased tissue from both humans and mice [13, 14].
Changes in epigenetic modifications and transcriptional regulation are essential for phenotypic adaptation of cell types to environments, including high-fat diet (HFD) [15,16,17]. Since some DNA-binding proteins play a role in triggering epigenetic reprogramming in specific genomic regions [18], we hypothesised that the corresponding sequence motifs could be enriched in cis-regulatory regions with local epigenetic alterations in response to environmental stimuli. In the present study, by examining genome-wide H3K27ac signatures and whole transcriptome, we aimed to identify sequence motifs enriched in genomic regions exhibiting HFD-driven epigenetic changes in mouse pancreatic islets. In addition, we investigated the involvement of DNA-binding proteins in the regulation of gene expression and pancreatic beta cell function during the development of obesity and glucose intolerance.
Methods
Mice and metabolic assessment
Male C57BL/6JJcl mice were purchased from CLEA Japan (Tokyo, Japan). Half of the mice were switched from a standard chow diet (CE-2; CLEA Japan) to an HFD (D12492, 60% fat; Research Diets, New Brunswick, NJ, USA) at 10 weeks of age. The samples were not randomised by statistical computing. Mice were weighed to the nearest 0.1 g. Blood glucose was determined by tail-vein bleeding. Following fasting of the mice overnight, an intraperitoneal glucose tolerance test (IPGTT) was conducted using a glucose dose of 1 g/kg body weight. Plasma was separated by centrifugation of whole blood at an ambient temperature of 22 ± 2°C and frozen in liquid nitrogen for measurement of insulin levels using a Mouse Insulin ELISA Kit (Morinaga Institute of Biological Science, Kanagawa, Japan). All animal procedures at the National Center for Global Health and Medicine were approved by the Institutional Animal Care and Use Committee. Blinding for experimenters was not carried out. We did not lay down any criteria for inclusion and exclusion of any data, since all the results supported the main findings of the study. For details, see electronic supplementary material (ESM) Methods.
Isolation of pancreatic islets
Pancreatic islets were isolated from anaesthetised 37-week-old mice as described [19]. See ESM Methods for details.
High-throughput RNA sequencing
Total RNA was extracted from pancreatic islets using Trizol (Thermo Fisher Scientific, Waltham, MA, USA). Library preparation was performed using the TruSeq Stranded Total RNA Sample Prep Kit (Illumina, San Diego, CA, USA) and high-throughput RNA sequencing (RNA-Seq) was performed using the Illumina HiSeq 2000 platform with 100 bp paired-end sequencing. See ESM Methods for details.
Analysis of differential gene expression and functional annotation
Analyses were performed using DESeq 1.32.0 [20] with an iterative pipeline (iterative differentially expressed gene elimination strategy [iDEGES]/DESeq) [21], both of which were R/Bioconductor packages (https://www.bioconductor.org/). Reads per million (RPM)-normalised read counts allowed the visualisation of global changes in gene expression as an MA plot. For those genes with highest variance (p < 0.1), functional annotation was analysed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) 6.8 (https://david.ncifcrf.gov/, accessed 15 Feb 2018) [22]. See ESM Methods for details.
Chromatin immunoprecipitation, high-throughput sequencing and analysis of differential sequencing signals
Chromatin preparation for chromatin immunoprecipitation (ChIP) was performed as described in [4] with modifications. A list of the primary antibodies used is provided in ESM Table 1. ChIP libraries were prepared using NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (New England BioLabs, Ipswich, MA, USA) and were sequenced on a HiSeq 2000 system (Illumina). The processed data were uploaded to the Integrative Genomics Viewer (IGV) browser 2.3.75 (http://software.broadinstitute.org/software/igv/) [23]. Spatial clustering for identification of ChIP-enriched regions (SICER) v1.1 [24] was used to compare differentially enriched regions between experimental and control groups using the command SICER-df-rb.sh. Running this command enabled identification of significantly enriched H3K27ac regions in each library, merging of all overlapping intervals and determination of the significance of changes by comparing the signal intensity of ChIP coupled with high-throughput sequencing (ChIP-Seq) in HFD samples with that in controls on each merged H3K27ac region. To identify DNA sequence motifs enriched in merged H3K27ac regions with HFD-induced changes, we used Hypergeometric Optimization of Motif EnRichment (HOMER) v4.6 (http://homer.ucsd.edu/homer/) [25] for de novo motif detection using the whole genome as the background by running the command findMotifsGenome.pl. This command is a tool used to identify binding motifs enriched in the genomic intervals without prior knowledge of the transcription factor binding sites. First, analyses were performed according to the default parameters for base redundancy and motif length (8, 10 or 12 bp). Subsequently, the identified motifs were checked for homology with known motif matrices and the best match result was presented as a likely candidate, which we listed in our tables. See ESM Methods for details.
Correlating differential H3K27ac signals with differential gene expression
Each H3K27ac region was mapped to the closest gene based on distance to the transcription start site (TSS), as described [14]. Overall cis-regulatory activity of individual genes was defined as increased, decreased or unchanged by counting the number of regions showing increased H3K27ac relative to regions showing a decrease. DESeq2 1.20.0 [26] enabled comparison of log2 (fold change) between categories. For Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis [27], we selected the 610 most upregulated and 174 most downregulated genes, which also showed overall increases and decreases in cis-regulatory activity, respectively. See ESM Methods for details.
Cell culture and insulin secretion measurements in INS-1 cells and pancreatic islets
INS-1 cells (a rat beta cell line) were gift from C. B. Wollheim (Lund University; University of Geneva) and N. Sekine (University of Geneva), and were cultured as described [19]. Batch insulin release was estimated by incubating in 3 mmol/l glucose, 25 mmol/l glucose or 30 mmol/l KCl for 60 min. For pancreatic islets isolated from 54-week-old mice, insulin secretion in 2.8 mmol/l glucose, 16.7 mmol/l glucose or 60 mmol/l KCl in 30 min batch incubations was measured as described [19]. Total insulin content was calculated by summating the insulin secreted into the medium with the amount of insulin measured by whole-cell extraction with acid ethanol for 24 h at 4°C. Insulin secretion was expressed as a percentage of the total insulin content. For INS-1 cells, insulin was detected using a Mouse Insulin ELISA Kit (AKRIN-011 T; Shibayagi, Gunma, Japan). Mycoplasma contamination was not tested. See ESM Methods for details.
siRNA knockdown of NRF1, GABPA and MEF2A followed by palmitate treatment
Lipofectamine 2000 (Thermo Fisher Scientific) was used for reverse transfection of specific siRNAs in INS-1 cells (ESM Table 2). Palmitate (Wako Pure Chemical Industries, Osaka, Japan), sodium palmitate (Sigma-Aldrich, St Louis, MO, USA) or vehicle treatment began 36 h after siRNA-mediated knockdown, and treated cells were used for the following experiments after incubation for 48 h. See ESM Methods for details.
Quantitative real-time PCR
Quantification of cDNA or ChIP-DNA samples was conducted via standard quantitative PCR (qPCR) methods using SYBR Green (Thermo Fisher Scientific). The following genes were tested: Acaa2, Gabpa, Mef2a, Nrf1 and Slc25a20 for rat cDNA; Acaa2, Cpt1a, Elovl6, Fads2, Mki67, Scd2, Slc25a20 and Top2a for mouse cDNA and Acaa2 and Slc25a20 for rat ChIP-DNA. A list of primers is provided in ESM Table 3. See ESM Methods for details.
Statistics
We used R or GraphPad Prism 7.04 (GraphPad Software, San Diego, CA, USA) for statistical analyses. The two-sided unpaired t tests, two-sided paired t tests, or Mann–Whitney U tests were applied. Error bars in the Figures represent SD.
Accession numbers
Previous ChIP-Seq data in human or mouse tissues were obtained from Gene Expression Omnibus (GEO) and processed as described above (summarised in ESM Table 4). The UCSC liftOver tool and Ensembl Compara cross-species resources (https://www.ensembl.org/info/genome/compara/index.html) were applied for conversion of genomic positions between reference genomes mm9, hg19 and rn6. Data in the present study were deposited at ArrayExpress under accession number E-MTAB-6718 (RNA-Seq) and E-MTAB-6719 (ChIP-Seq).
Results
Body weight and glucose tolerance in mouse model of diet-induced obesity
To investigate the mechanisms by which environmental factors induce phenotypic differences in pancreatic beta cells, a diet-induced obesity (DIO) model was employed, using male C57BL/6J mice [28]. The HFD was started at 10 weeks of age (Fig. 1a). The initial 4 weeks of HFD feeding resulted in a slight but significant increase in average body weight (control mice 27.0 ± 2.05 g vs DIO mice 29.2 ± 3.04 g, p < 0.01, ESM Fig. 1a) and non-fasting blood glucose level (control mice 9.19 ± 1.6 mmol/l vs DIO mice 10.1 ± 1.6 mmol/l, p < 0.05, ESM Fig. 1b). IPGTT showed that glucose tolerance was impaired in DIO mice, although these mice displayed enhanced glucose-stimulated insulin secretion (GSIS) at 15 min post-glucose injection (ESM Fig. 1c,d). Thereafter, mildly higher non-fasting blood glucose levels were sustained in DIO mice for 37 weeks of HFD feeding (total age 47 weeks; Fig. 1a–c). During IPGTT, DIO mice showed mild glucose intolerance (Fig. 1d) and hyperinsulinaemia both at baseline (8.8-fold, p < 0.01; Fig. 1e) and at 15 min post-glucose injection (7.5-fold, p < 0.01; Fig. 1e).
Generation and characterisation of DIO mice. (a) The time course diagram and analytical strategies. (b–e) Metabolic assessment of 47-week-old mice (after completing 37 weeks of HFD feeding), showing body weight (b; n = 29 for control group and n = 22 for DIO group), non-fasting blood glucose (c; n = 14 for control group and n = 10 for DIO group), IPGTT glucose excursion (d; grey circles, control, n = 4; orange circles, DIO, n = 4) and IPGTT plasma insulin values (e; grey bars, control, n = 4; orange bars, DIO, n = 4). Data are means ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001 (in d, DIO vs control at specific timepoint)
Impact of long-term HFD feeding on pancreatic islet transcriptome
To clarify the global relationships between epigenome and transcriptome in the pancreatic islets after HFD feeding for an extended period of 27 weeks, we used a next-generation sequencing platform (Fig. 1a and ESM Fig. 2). RNA-Seq analyses were performed by generating approximately 31.7–35.1 million high-quality paired-end sequencing reads (ESM Table 5).
To identify genes differentially expressed between DIO and control mice, we chose DESeq [20] with an iterative pipeline [21] for read count normalisation and gene ranking. The analysis revealed that most genes showed relatively limited dynamics with respect to the read count ratio (ESM Fig. 3a). Nevertheless, analysis of the top-ranked 303 upregulated genes (p < 0.1) (ESM Fig. 3b and ESM Tables 6, 7) using DAVID [22] revealed enrichment of terms that were linked to islet functions important for adaptive responses to HFD, including the endoplasmic reticulum [29] (ESM Fig. 3b and ESM Table 7).
We also performed the same analysis on the top-ranked 269 downregulated genes (p < 0.1) (ESM Fig. 3c and ESM Tables 6, 8). Interestingly, the enriched terms were mainly related to epigenetic regulation of gene transcription, which may suggest the involvement of epigenetic change in pancreatic islets following prolonged HFD feeding (ESM Fig. 3c and ESM Table 8).
Genome-wide changes in H3K27ac associated with differential gene expression under HFD feeding
H3K27ac ChIP-Seq was used for genome-wide identification of active cis-regulatory elements in the pancreatic islets (ESM Fig. 2). Approximately 88% of 43.8–49.6 million reads were aligned to the mouse reference genome (ESM Table 9). Using SICER [24], we determined 37,534 and 40,207 H3K27ac-enriched regions genome-wide for control and DIO libraries with median size of 3.6 kb and 3.2 kb (ESM Tables 10, 11), respectively, further generating a catalogue of 39,350 merged regions as putative cis-regulatory regions shared between our samples with median size of 3.8 kb (Fig. 2a, ESM Fig. 2 and ESM Table 12).
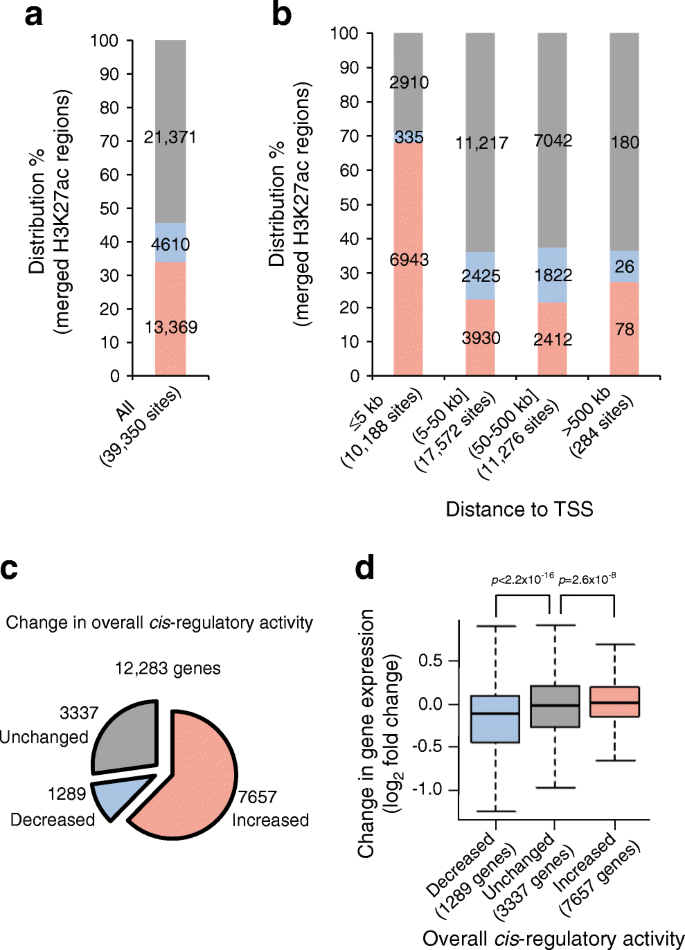
RNA-Seq and H3K27ac ChIP-Seq analysis of pancreatic islets derived from DIO mice. (a) Bar plot showing the number of merged H3K27ac regions that were increased, decreased or unchanged upon HFD feeding. (b) Genomic distribution of H3K27ac regions showing HFD-induced changes in ChIP-Seq signal intensity for regions in ≤5 kb, 5–50 kb, 50–500 kb and > 500 kb distance from TSSs. Pink bars, increased H3K27ac sites; blue bars, decreased H3K27ac sites; grey bars, unchanged H3K27ac sites. (c) Pie chart indicating the number of genes associated with cis-regulatory regions, where the direction of change in cis-regulatory activity as a whole was regarded to be increased, decreased or unchanged. Only genes that could be tested for differential gene expression by DESeq were included. Data without knowledge of DESeq results are shown in ESM Fig. 5b. (d) Box plot showing differential expression of genes associated with cis-regulatory elements, the overall activity of which was decreased, unchanged and increased. The horizontal line inside the box is the median value. The upper and lower ends of the box are the 75th and 25th percentile for values, respectively. The vertical lines from the ends of the box (whiskers) represent the smallest or largest observed values within 1.5 times the interquartile range. p values were calculated using the Mann–Whitney U test
SICER also detected changes in the ChIP-Seq signal in individual merged regions [24]. We found 13,369 increased and 4610 decreased H3K27ac regions in the islets derived from DIO mice (Fig. 2a, ESM Fig. 2 and ESM Table 12), with median fold changes in normalised enrichment of 1.30 (first quartile 1.20, third quartile 1.41) and 0.73 (first quartile 0.67, third quartile 0.80), respectively. Compared with unchanged and decreased H3K27ac, we found that increased H3K27ac showed a greater distribution in the promoter-proximal regions (Fig. 2b and ESM Fig. 4a–d). Of all the H3K27ac regions, increased H3K27ac accounted for 68.1% (6943/10,188) from −5 kb to +5 kb relative to the TSS (Fig. 2b), while it accounted for less than 22.0% (6420/29,132) if located >5 kb relative to the TSS (Fig. 2b).
To connect epigenetic changes with the transcriptome, a precise regulatory map of the cell types is vital [30]. In the present study, however, we assigned a single gene to each merged H3K27ac region using only genomic positioning information (termed a ‘single nearest gene association rule’ in the Genomic Regions Enrichment of Annotations Tool [GREAT] [31]). A similar approach was also employed in a previous study that successfully analysed a mouse model of Alzheimer’s disease [14]. Accordingly, expression of 9391 and 2794 genes were assumed to be associated with increased and decreased H3K27ac, respectively (ESM Table 6 and ESM Fig. 5a).
As 13.3% (1632/12,283) of all H3K27ac-harbouring genes revealed regulatory regions consisting of both increased and decreased H3K27ac (ESM Fig. 5c and ESM Table 6), we investigated how changes in individual gene expression arose according to the number of increased H3K27ac sites relative to those decreased (Fig. 2c,d and ESM Fig. 5b). Compared with genes containing equal numbers of increased and decreased H3K27ac regions, the median log2 fold change in gene expression was significantly larger when the increased H3K27ac was superior in number in the cis-regulatory domain (p = 2.6 × 10−8; Fig. 2d), whereas it was significantly smaller when the number of increased H3K27ac regions was less than the decreased ones (p < 2.2 × 10−16; Fig. 2d).
De novo motif analysis of HFD-responsive H3K27ac regions revealed enrichment of novel DNA-binding sites
We sought to determine sequence motifs for DNA-binding proteins that were significantly enriched in differentially enriched H3K27ac regions by HOMER [25], the de novo motif analysis (ESM Fig. 2). Increased H3K27ac in the islets of DIO mice revealed a significant enrichment for 12 motifs (threshold, p < 1 × 10−12; Table 1), including binding motifs for known transcription factors, such as regulatory factor X, 7 (RFXDC2) [32], forkhead box A1 (FOXA1) [33] and HNF1 homeobox A (HNF1) [34]. However, the analysis also revealed highly enriched motifs exhibiting a close match to the consensus sequences for nuclear respiratory factor 1 (NRF1), GA repeat binding protein α (GABPA) and myocyte enhancer factor 2A (MEF2A) (all with score > 0.900; Table 1), the biological relevance of which had not been well described for beta cells. Further, these motifs were not detected either in unchanged or decreased H3K27ac regions (ESM Tables 13 and 14). We found that there were 3168 NRF1, 3281 GABPA and 1366 MEF2A motifs in 2369, 2555 and 1207 increased H3K27ac regions, respectively. These regions were further connected to 2318, 2497 and 1113 putative target genes, respectively (Table 1). We found only one motif for v-maf musculoaponeurotic fibrosarcoma oncogene family protein K (MAFK) in decreased H3K27ac, with moderate significance (score = 0.617, p = 1 × 10−14; ESM Table 13).
NRF1 and GABPA are involved in local epigenomic regulation in INS-1 cells of the palmitate-induced expression of Acaa2 and Slc25a20 for β-oxidation
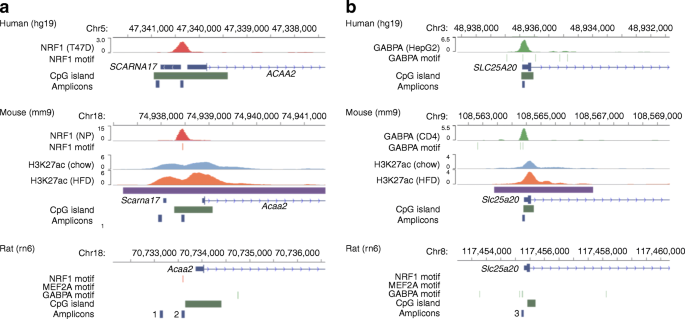
Next, we tested how these transcription factors could be involved in the regulation of H3K27ac and gene expression, especially under an environmental condition mimicking DIO mice. Within the HFD model, the exogenous factors triggering epigenomic modifications to specific genomic loci in beta cells are very important. There are many possible candidates but among them we examined the role of NEFA using INS-1 cells, a rat beta cell line. Since our data suggested that the regulatory effect of H3K27ac was not always strong (Fig. 2d), we first selected the most variable 1000 genes that were either upregulated or downregulated (Fig. 3). Among them, we found that 610 upregulated and 174 downregulated genes also exhibited a change in the overall H3K27ac in the same direction (ESM Fig. 6a,b). To obtain functional insights into these distinct gene sets, we performed KEGG-based pathway analysis [27]. Among various enriched metabolic pathways (ESM Tables 15, 16), those related to fatty acid metabolism (degradation and biosynthesis of fatty acids) were identified both in upregulated and downregulated genes, respectively (Fig. 3, Table 2 and ESM Tables 15, 16). These findings prompted us to focus on two genes, Acaa2 and Slc25a20; both are involved in mitochondrial fatty acid β-oxidation and have been reported to be upregulated in the presence of high NEFA levels [35,36,37]. Furthermore, previous ChIP-Seq data suggested that binding of NRF1 and GABPA to their binding motifs was located close to the TSSs of Acaa2 and Slc25a20 (Fig. 4a,b, ESM Fig. 7), respectively.
MA plot for differential analysis of gene expression between DIO and control samples using DESeq. The plots for selected genes are represented in red or dark blue. The light red or light blue represent the 1000 top-ranked genes that exhibited upregulation or downregulation, respectively, facilitating the identification of genes with higher variance. A zoomed image is also presented
Profiles of islet H3K27ac in the Acaa2 and Slc25a20 loci and the inference of NRF1 and GABPA binding sites, respectively, from previously deposited ChIP-Seq data. H3K27ac ChIP-Seq profiles are shown for control (light blue) and experimental DIO (orange) mice; purple bars track H3K27ac increases in the experimental data. Previously deposited ChIP-Seq data for NRF1 (red) or GABPA (light green) are also included, together with vertical lines for the DNA-binding motifs. CpG islands and qPCR products are represented by dark green and dark blue bars, respectively. To show the evolutionary conservation of the locus, panels for human (hg19), mouse (mm9) and rat (rn6) are arranged vertically. Scale bars represent the coverage expressed as read counts normalised per million reads. For further information, including the results of other cell types and MEF2A ChIP-Seq, see ESM Fig. 7. (a) The Acaa2 locus showed an increase in H3K27ac surrounding the TSS, which was HFD responsive. A conserved NRF1 motif was found within this region in both mice and rats but not in humans, due to a single base-pair substitution. Mice showed a strong and localised binding peak of NRF1 surrounding the motif. Two primer sets, amplicon 1 and amplicon 2, for the rat genome were designed to detect H3K27ac enrichment and NRF1 binding, respectively, as shown in the track labelled ‘Amplicons’. Conversion of genomic intervals of these amplicons was conducted using the UCSC liftOver tool to identify orthologous regions in the panels for mouse (mm9) and human (hg19). (b) The Slc25a20 locus showed the increase in H3K27ac surrounding the TSS. Within this region, GABPA showed a localised binding to its motif both in humans and mice. Amplicon 3 was designed to detect GABPA binding and H3K27ac enrichment in the 5′ region in INS-1 cells, for which orthologous regions are indicated in the panels for mm9 and hg19. CD4, mouse CD4 T cells; Chr, chromosome; HepG2, Hep G2 human hepatoblastoma cell line; NP, neuronal progenitors; T47D, T47D human ductal breast cancer cell line
We investigated the effects of 0.5 mmol/l palmitate on the epigenome, as well as the expression of Acaa2 or Slc25a20 in INS-1 cells, in the presence or absence of the siRNA-mediated knockdown of either NRF1 or GABPA (Fig. 5). Nrf1 siRNA and Gabpa siRNA treatment reduced mRNA levels by 35% (p < 0.001; Fig. 5a) and 46% (p < 0.001; Fig. 5e), respectively. At 48 h after palmitate treatment, we found that mRNA levels of Acaa2 and Slc25a20 in INS-1 cells increased approximately 6.1-fold (p < 0.01; Fig. 5b) and 2.6-fold (p < 0.001; Fig. 5f) compared with the vehicle-treated control, respectively. Although ChIP-qPCR revealed NRF1 and GABPA binding to expected sites containing their binding motifs (Fig. 4a,b and ESM Fig. 8a,c), enhanced binding was not observed after palmitate treatment (Fig. 5c,g and ESM Fig. 8a,c). However, suppression of binding for each of these transcription factors by siRNA-mediated knockdown resulted in significant downregulation of both Acaa2 (1.95-fold, p < 0.001; Fig. 5b) and Slc25a20 (1.45-fold, p < 0.05; Fig. 5f), indicating that they were direct targets of NRF1 and GABPA, respectively. Although palmitate allowed a similar increase in mRNA levels under knockdown conditions (sevenfold increase in Acaa2 under NRF1 knockdown, p < 0.001, Fig. 5b; 2.9-fold increase in Slc25a20 under GABPA knockdown, p < 0.01, Fig. 5f), the resultant mRNA levels were about 59.2% (p < 0.01; Fig. 5b) and 77.7% (p < 0.05; Fig. 5f) of the values observed when the INS-1 cells were treated with both scrambled siRNA and palmitate. Thus, NRF1 and GABPA were involved in the expression of Acaa2 and Slc25a20, respectively, both at baseline and following palmitate treatment.
Roles of NRF1, GABPA and MEF2A in gene expression and H3K27ac enrichment in basal and palmitate-treated INS-1 cells. Cells were initially treated with scrambled siRNA, Nrf1 siRNA, Gabpa siRNA or Mef2a siRNA and treated 36 h later with vehicle or 0.5 mmol/l palmitate. (a–d) Nrf1 siRNA treatment. Relative mRNA levels of Nrf1 (a) and Acaa2 (b) were measured after Nrf1 siRNA treatment. Cells were subjected to ChIP using an anti-NRF1 antibody or IgG (control) (c). ChIP-enriched DNA was quantified by qPCR using primer pairs to detect NRF1 binding to Acaa2 (amplicon 2 in Fig. 4) and Nanog promoter (negative control). Values are expressed as a percentage of input DNA. The displayed data show only results of amplicon 2 for NRF1 ChIP. H3K27ac ChIP-qPCR analysis was carried out (d). Enrichment within Acaa2 promoter (amplicon 1 in Fig. 4) and Nanog promoter (negative control) is expressed as a percentage of input DNA. The displayed data show only results of amplicon 1 for H3K27ac ChIP. (e–h) Gabpa siRNA treatment. Relative mRNA levels of Gabpa (e) and Slc25a20 (f) were measured after Gabpa siRNA treatment. Cells were subjected to ChIP using an anti-GABPA antibody or IgG (control) (g). ChIP-enriched DNA was quantified by qPCR with primer pairs to detect GABPA binding to Slc25a20 (amplicon 3 in Fig. 4) and Nanog promoter (negative control). Values are expressed as a percentage of input DNA. The displayed data show only results of amplicon 3 for GABPA ChIP. H3K27ac ChIP-qPCR analysis was carried out (h). Enrichment within Slc25a20 promoter (amplicon 3 in Fig. 4) and Nanog promoter (negative control) was expressed as a percentage of input DNA. The displayed data show only results of amplicon 3 for H3K27ac ChIP. (i–k) Mef2a siRNA treatment. Relative mRNA levels of Mef2a (i), Acaa2 (j) and Slc25a20 (k) were measured after Mef2a siRNA treatment. For all ChIP-qPCR analyses, all data, including those for IgG and Nanog promoter, are shown in ESM Fig. 8. Normalised mRNA levels were calculated relative to baseline, where Actb was used as an internal control. Data are shown as means ± SD, n = 3 (except for NRF1 ChIP of Nrf1 siRNA and palmitate-treated samples, where n = 2.). *p < 0.05, **p < 0.01 and ***p < 0.001 compared with baseline or for comparisons indicated by horizontal line (calculated by two-sided unpaired t test)
For epigenetic analysis, we measured H3K27ac enrichment using qPCR primer sets for cis-regulatory elements of interest (Fig. 4, ESM Fig. 7 and ESM Table 3). Within the regions corresponding to the islet-merged H3K27ac regions spanning the TSS, H3K27ac enrichment in Acaa2 and Slc25a20 was lower than the control by 37.6% (p < 0.05; Fig. 5d and ESM Fig. 8b) and 28.8% (p < 0.05; Fig. 5h and ESM Fig. 8d) upon knockdown of NRF1 and GABPA, respectively. As observed in quantitative RT-PCR analysis of Acaa2 or Slc25a20 transcripts (Fig. 5b,f), ChIP-qPCR showed that the increase in H3K27ac in response to palmitate was blunted by knockdown of NRF1 and GABPA, respectively (Fig. 5d,h and ESM Fig. 8b,d), although the fold change in H3K27ac enrichment before and after palmitate treatment was similar regardless of whether NRF1 or GABPA was suppressed by siRNA. H3K27ac of Acaa2 increased in scrambled-siRNA-treated cells by 2.43-fold (p < 0.01), whereas it increased in Nrf1-siRNA-treated cells by 2.57-fold (p < 0.01) (Fig. 5d and ESM Fig. 8b). Although palmitate increased H3K27ac in Slc25a20 by 1.46-fold in control cells (p < 0.05; Fig. 5h and ESM Fig. 8d), a significant increase was not observed when GABPA was suppressed (Fig. 5h and ESM Fig. 8d).
MEF2A regulated fatty acid β-oxidation genes, Acaa2 and Slc25a20, similarly to NRF1 and GABPA
Further, we tested whether MEF2A regulated Acaa2 and Slc25a20. Rather than a localised peak, the TSS regions of Acaa2 and Slc25a20 genes revealed modest and relatively broad signals in previously deposited MEF2A ChIP-Seq data (ESM Fig. 7). Treatment of INS-1 cells with Mef2a siRNA resulted in a reduction in mRNA level of 18.8% (p < 0.01; Fig. 5i). The downregulation of Acaa2 and Slc25a20 was not significant following MEF2A knockdown (Fig. 5j,k). However, MEF2A knockdown resulted in 26.7% (p < 0.05) and 30.1% (p < 0.01) decrease in expression of Acaa2 and Slc25a20, respectively, in the presence of palmitate treatment (Fig. 5j,k). Taken together, these results suggest the important roles of the transcription factor motifs and their binding transcription factors in the regulation of genes and epigenome for fatty acid β-oxidation.
Knockdown of binding transcription factors for enriched motifs resulted in increased basal insulin secretion in INS-1 cells
To gain insight into the functional effects of increased H3K27ac containing the above-described motifs on beta cells, we performed GSIS tests in INS-1 cells treated with Nrf1, Mef2a or Gabpa siRNA. For Nrf1, besides cells treated with siRNA and vehicle, we also tested GSIS for knockdown cells that were treated with 0.5 mmol/l palmitate for 48 h to observe the effect of environmental load on cells with impaired regulation of motif-mediated transcription (Fig. 6a,d). Compared with control cells treated with scrambled siRNA, incubation in 3 mmol/l glucose resulted in increases in basal insulin secretion of 1.82-fold (p < 0.001; Fig. 6a), 3.09-fold (p < 0.001; Fig. 6b) and 2.35-fold (p < 0.001; Fig. 6c) in NRF1, MEF2A and GABPA knockdown cells, respectively. Another remarkable effect of the knockdown of these transcription factors was the reduction in insulin content at 3 mmol/l glucose, which was observed for the knockdown of MEF2A (33.9%; p < 0.05; Fig. 6e) and GABPA (26.6%; p < 0.01; Fig. 6f). In response to stimulation with 25 mmol/l glucose or 30 mmol/l KCl, there was no change in the secretory response of either MEF2A or GABPA knockdown cells (Fig. 6b,c), whereas 25 mmol/l glucose or 30 mmol/l KCl, respectively, caused a 14.0% (p < 0.05; Fig. 6a) increase and a 23.4% (p < 0.01; Fig. 6a) decrease in insulin secretion in NRF1 knockdown cells. In the present study, 48 h treatment with 0.5 mmol/l palmitate resulted in a significant increase in insulin secretion following incubation in 3 mmol/l glucose (1.44-fold, p < 0.05; Fig. 6a), along with a significant decrease in insulin content (35.6% reduction, p < 0.05; Fig. 6d), similar to the findings in the knockdown experiments. Furthermore, we observed a marked increase in both basal insulin secretion (3.36-fold; p < 0.01; Fig. 6a) and insulin response to 25 mmol/l glucose (1.36-fold, p < 0.01; Fig. 6a) in NRF1 knockdown cells treated with 0.5 mmol/l palmitate.
GSIS and insulin content of INS-1 cells after siRNA-mediated knockdown of NRF1, MEF2A or GABPA. (a) GSIS in INS-1 cells after transfection of Nrf1 siRNA; 0.5 mmol/l sodium palmitate was added 36 h after transfection. White bars, scrambled siRNA; pink bars, scrambled siRNA and palmitate; blue bars, Nrf1 siRNA; orange bars, Nrf1 siRNA and palmitate. (b) GSIS in INS-1 cells 72 h after transfection of Mef2a siRNA. White bars, scrambled siRNA; blue bars, Mef2a siRNA. (c) GSIS in INS-1 cells 72 h after transfection of Gabpa siRNA. White bars, scrambled siRNA; blue bars, Gabpa siRNA. (d) Total insulin content in INS-1 cells after transfection of Nrf1 siRNA; 0.5 mmol/l sodium palmitate was added 36 h after transfection. White bars, scrambled siRNA; pink bars, scrambled siRNA and palmitate; blue bars, Nrf1 siRNA; orange bars, Nrf1 siRNA and palmitate. (e) Total insulin content in INS-1 cells after transfection of Mef2a siRNA. White bars, scrambled siRNA; blue bars, Mef2a siRNA. (f) Total insulin content in INS-1 cells after transfection of Gabpa siRNA. White bars, scrambled siRNA; blue bars, Gabpa siRNA. For each well, INS-1 cells were incubated in the indicated testing solution for 1 h to measure insulin secretion. Palmitate treatment was conducted for 48 h. n = 4. *p < 0.05, **p < 0.01 and ***p < 0.001 compared with baseline or for comparisons indicated by horizontal line (calculated by two-sided unpaired t test)
Discussion
In the present study, we show that the histone modification H3K27ac characteristic of active cis-regulatory regions changes purely through environmental effects by comparing samples derived from genetically identical C57BL/6J mice fed an HFD. Despite heterogeneity in the cellular composition, the global gene expression profile in pancreatic islets can be largely explained by that in beta cells [10], thus facilitating the analysis of whole islets and further examination of molecular mechanisms using INS-1 cells.
Previous large-scale analyses have demonstrated the correlation between changes in H3K27ac and gene expression [14]. Although transcription factors serve as direct transcriptional activators [38], they may also be involved in inducing epigenetic alterations in specific regions of the genome [18]. Consistent with this, our analyses revealed that environmental changes resulted in genome-wide epigenetic alterations, which correlated with differential gene expression. Furthermore, by de novo motif analysis, we identified enrichment of sequence motifs in H3K27ac regions showing changes in response to HFD, among which transcription factors such as NRF1, GABPA and MEF2A were the most enriched binding motifs. This led to the identification of potential target genes, Acaa2 and Slc25a20, which are important for fatty acid β-oxidation. Our in vitro analysis using INS-1 cells produced results consistent with such in vivo findings. Suppression of NRF1, GABPA or MEF2A by siRNA resulted in blunted responses of both H3K27ac gain surrounding the TSS and gene induction to palmitate treatment, especially in the case of genes involved in fatty acid metabolism. Therefore, the environmental factors, including fat-rich diet, shaped epigenetically induced specific pathways that were generated by the contribution of specific transcription factors.
NRF1, GABPA and MEF2A are transcription factors that have been linked to mitochondrial regulation [39]; however, their molecular function in pancreatic beta cells has not been well characterised. The knockdown experiments using INS-1 cells showed that changes in the secretory profile occurred in response to different testing solutions. This was in marked contrast to the reduced in vitro glucose responsiveness of pancreatic islets derived from 54-week-old hyperinsulinaemic DIO mice with mild glucose intolerance (ESM Fig. 9), in which the gene expression profile was similar to that of 37-week-old DIO mice (ESM Fig. 10). The epigenomic state should reflect the increased transcription factor activity (Table 1); thus, it could be possible that changing NRF1 activity could be useful in modulating pancreatic beta cell function. Besides this, the importance of the NRF1 gene was shown by a recent GWAS demonstrating the association of an NRF1 locus to the childhood obesity-related trait [40].
We also identified enrichment of a binding motif for MAFK in decreased H3K27ac regions. MAFK has been reported to be a negative regulator of pancreatic beta cell function and inhibition of endogenous small-MAFs improved glucose tolerance in DIO mice [41]. Therefore, our findings of decreased H3K27ac regions could be responsible, at least in part, for mild glucose intolerance in DIO mice.
Our study supports the potential use of epigenetic analyses, such as H3K27ac, in a wide variety of applications for the research of human complex diseases. In this respect, the application of transcription factor-mediated repression of the epigenome has potential as an effective intervention against obesity and type 2 diabetes in the future.
Data availability
The next-generation sequencing data in the present study were deposited at ArrayExpress.
RNA-Seq:
Dataset name: ERR2538129 (Control), ERR2538130 (Diet-induced obesity)
Repository name and number: E-MTAB-6718 - RNA-Seq of pancreatic islets derived from mice fed a long-term high-fat diet against chow-fed controls.
ChIP-Seq:
Dataset name: ERR2538131 (Control), ERR2538132 (Diet-induced obesity)
Repository name and number: E-MTAB-6719 - H3K27ac ChIP-Seq of pancreatic islets derived from mice fed a long-term high-fat diet (HFD) against chow-fed controls.
Abbreviations
- ChIP:
-
Chromatin immunoprecipitation
- ChIP-Seq:
-
Chromatin immunoprecipitation coupled with high-throughput sequencing
- DAVID:
-
The Database for Annotation, Visualization and Integrated Discovery
- DIO:
-
Diet-induced obesity
- FOXA1:
-
Forkhead box A1
- GABPA:
-
GA repeat binding protein α
- GEO:
-
Gene Expression Omnibus
- GSIS:
-
Glucose-stimulated insulin secretion
- GWAS:
-
Genome-wide association study (studies)
- H3K27ac:
-
Histone H3 lysine 27 acetylation
- HFD:
-
High-fat diet
- HNF1:
-
HNF1 homeobox A
- HOMER:
-
Hypergeometric Optimization of Motif EnRichment
- IPGTT:
-
Intraperitoneal glucose tolerance test
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MAFK:
-
v-maf musculoaponeurotic fibrosarcoma oncogene family protein K
- MEF2A:
-
Myocyte enhancer factor 2A
- NRF1:
-
Nuclear respiratory factor 1
- qPCR:
-
Quantitative PCR
- RFXDC2:
-
Regulatory factor X, 7
- RNA-Seq:
-
High-throughput RNA sequencing
- SICER:
-
Spatial clustering for identification of ChIP-enriched regions
- TSS:
-
Transcription start site
References
Caspard H, Jabbour S, Hammar N, Fenici P, Sheehan JJ, Kosiborod M (2018) Recent trends in the prevalence of type 2 diabetes and the association with abdominal obesity lead to growing health disparities in the USA: an analysis of the NHANES surveys from 1999 to 2014. Diabetes Obes Metab 20(3):667–671. https://doi.org/10.1111/dom.13143
Flegal KM, Carroll MD, Ogden CL, Curtin LR (2010) Prevalence and trends in obesity among US adults, 1999-2008. JAMA 303(3):235–241. https://doi.org/10.1001/jama.2009.2014
Visscher PM, Wray NR, Zhang Q et al (2017) 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet 101(1):5–22. https://doi.org/10.1016/j.ajhg.2017.06.005
Gaulton KJ, Nammo T, Pasquali L et al (2010) A map of open chromatin in human pancreatic islets. Nat Genet 42(3):255–259. https://doi.org/10.1038/ng.530
Maurano MT, Humbert R, Rynes E et al (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337(6099):1190–1195. https://doi.org/10.1126/science.1222794
Farh KK, Marson A, Zhu J et al (2015) Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518(7539):337–343. https://doi.org/10.1038/nature13835
Kahn SE, Hull RL, Utzschneider KM (2006) Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444(7121):840–846. https://doi.org/10.1038/nature05482
Kulkarni RN, Stewart AF (2014) Summary of the Keystone islet workshop (April 2014): the increasing demand for human islet availability in diabetes research. Diabetes 63(12):3979–3981. https://doi.org/10.2337/db14-1303
Stitzel ML, Sethupathy P, Pearson DS et al (2010) Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab 12(5):443–455. https://doi.org/10.1016/j.cmet.2010.09.012
Nica AC, Ongen H, Irminger JC et al (2013) Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res 23(9):1554–1562. https://doi.org/10.1101/gr.150706.112
Pasquali L, Gaulton KJ, Rodríguez-Seguí SA et al (2014) Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 46(2):136–143. https://doi.org/10.1038/ng.2870
Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470(7333):279–283. https://doi.org/10.1038/nature09692
Cheng Y, Ma Z, Kim BH et al (2014) Principles of regulatory information conservation between mouse and human. Nature 515(7527):371–375. https://doi.org/10.1038/nature13985
Gjoneska E, Pfenning AR, Mathys H et al (2015) Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518(7539):365–369. https://doi.org/10.1038/nature14252
Carrer A, Parris JL, Trefely S et al (2017) Impact of a high-fat diet on tissue acyl-coA and histone acetylation levels. J Biol Chem 292(8):3312–3322. https://doi.org/10.1074/jbc.M116.750620
Malmgren S, Spégel P, Danielsson AP et al (2013) Coordinate changes in histone modifications, mRNA levels, and metabolite profiles in clonal INS-1 832/13 β-cells accompany functional adaptations to lipotoxicity. J Biol Chem 288(17):11973–11987. https://doi.org/10.1074/jbc.M112.422527
Siersbæk M, Varticovski L, Yang S et al (2017) High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss. Sci Rep 7(1):40220. https://doi.org/10.1038/srep40220
Zaret KS (2018) Pioneering the chromatin landscape. Nat Genet 50(2):167–169. https://doi.org/10.1038/s41588-017-0038-z
Nishimura W, Eto K, Miki A et al (2013) Quantitative assessment of Pdx1 promoter activity in vivo using a secreted luciferase reporter system. Endocrinology 154(11):4388–4395. https://doi.org/10.1210/en.2012-2248
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106. https://doi.org/10.1186/gb-2010-11-10-r106
Sun J, Nishiyama T, Shimizu K, Kadota K (2013) TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 14(1):219. https://doi.org/10.1186/1471-2105-14-219
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
Robinson JT, Thorvaldsdóttir H, Winckler W et al (2011) Integrative genomics viewer. Nat Biotechnol 29(1):24–26. https://doi.org/10.1038/nbt.1754
Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W (2009) A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25(15):1952–1958. https://doi.org/10.1093/bioinformatics/btp340
Heinz S, Benner C, Spann N et al (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38(4):576–589. https://doi.org/10.1016/j.molcel.2010.05.004
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27(1):29–34. https://doi.org/10.1093/nar/27.1.29
Clee SM, Attie AD (2007) The genetic landscape of type 2 diabetes in mice. Endocr Rev 28(1):48–83. https://doi.org/10.1210/er.2006-0035
Sharma RB, O’Donnell AC, Stamateris RE et al (2015) Insulin demand regulates β cell number via the unfolded protein response. J Clin Invest 125(10):3831–3846. https://doi.org/10.1172/JCI79264
Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295(5558):1306–1311. https://doi.org/10.1126/science.1067799
McLean CY, Bristor D, Hiller M et al (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28(5):495–501. https://doi.org/10.1038/nbt.1630
Smith SB, Qu HQ, Taleb N et al (2010) Rfx6 directs islet formation and insulin production in mice and humans. Nature 463(7282):775–780. https://doi.org/10.1038/nature08748
Gaulton KJ, Ferreira T, Lee Y et al (2015) Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet 47(12):1415–1425. https://doi.org/10.1038/ng.3437
Yamagata K, Oda N, Kaisaki PJ et al (1996) Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3). Nature 384(6608):455–458. https://doi.org/10.1038/384455a0
Busch AK, Gurisik E, Cordery DV et al (2005) Increased fatty acid desaturation and enhanced expression of stearoyl coenzyme A desaturase protects pancreatic β-cells from lipoapoptosis. Diabetes 54(10):2917–2924. https://doi.org/10.2337/diabetes.54.10.2917
Soni MS, Rabaglia ME, Bhatnagar S et al (2014) Downregulation of carnitine acyl-carnitine translocase by miRNAs 132 and 212 amplifies glucose-stimulated insulin secretion. Diabetes 63(11):3805–3814. https://doi.org/10.2337/db13-1677
Brun T, Scarcia P, Li N et al (2013) Changes in mitochondrial carriers exhibit stress-specific signatures in INS-1E β-cells exposed to glucose versus fatty acids. PLoS One 8(12):e82364. https://doi.org/10.1371/journal.pone.0082364
Lambert SA, Jolma A, Campitelli LF et al (2018) The human transcription factors. Cell 172(4):650–665. https://doi.org/10.1016/j.cell.2018.01.029
Scarpulla RC (2008) Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88(2):611–638. https://doi.org/10.1152/physrev.00025.2007
Comuzzie AG, Cole SA, Laston SL et al (2012) Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One 7(12):e51954. https://doi.org/10.1371/journal.pone.0051954
Nomoto H, Kondo T, Miyoshi H et al (2015) Inhibition of small Maf function in pancreatic β-cells improves glucose tolerance through the enhancement of insulin gene transcription and insulin secretion. Endocrinology 156(10):3570–3580. https://doi.org/10.1210/en.2014-1906
Acknowledgements
We thank C. B. Wollheim (Lund University, Lund, Sweden; University of Geneva, Geneva, Switzerland) and N. Sekine (University of Geneva) for providing INS-1 cells. We appreciate the assistance given by D. Suzuki, K. Nagase, N. Ishibashi (Lab Managers), H. Shiina and T. Shibuya (Administrative Assistants) (Department of Metabolic Disorder, National Center for Global Health and Medicine). We would like to thank Editage (www.editage.jp) for English language editing.
Contribution statement
TN and KY conceived this study. TN and HU performed the experiments. TN performed the computational analyses. TN and KY wrote the manuscript. TN, HU, NF, MK, TU, MH, WN and KY analysed the data, interpreted the results and contributed to discussions. The manuscript was critically reviewed, revised and given final approval by all co-authors. TN and KY are the guarantors of this work.
Funding
This work was supported by Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI), a grant from the National Center for Global Health and Medicine, the Japan Diabetes Foundation (to TN) and JSPS KAKENHI and a grant from the National Center for Global Health and Medicine (to KY). The study sponsors were not involved in the design of the study; the collection, analysis, and interpretation of data; writing the report; or the decision to submit the report for publication.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that there is no duality of interest associated with this manuscript.
Electronic supplementary material
ESM
(PDF 0.98 mb)
ESM Additional tables
(XLSX 5749 kb)
Rights and permissions
About this article
Cite this article
Nammo, T., Udagawa, H., Funahashi, N. et al. Genome-wide profiling of histone H3K27 acetylation featured fatty acid signalling in pancreatic beta cells in diet-induced obesity in mice. Diabetologia 61, 2608–2620 (2018). https://doi.org/10.1007/s00125-018-4735-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-018-4735-7