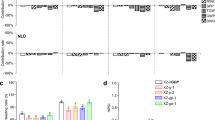
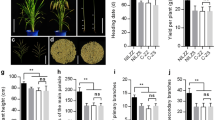
Abstract
Key message
We demonstrated the effect of heading time genes on reproductive transition and yield components under an identical genetic background using CRISPR/Cas9 gene-editing technology, and we propose that the elite allele will provide a new breeding strategy for rice breeding in high-latitude regions.
Abstract
Heading date is a factor closely associated with grain yield in rice (Oryza sativa L). In recent decades, a number of genes responsible for heading time have been identified, the variation of which contributes to the expansion of the rice cultivation area. However, it is difficult to compare the phenotypic effects of these genes due to the different genetic backgrounds. In this study, we generated 14 heading time mutants using CRISPR/Cas9 gene-editing technology and marker-assisted selection with a japonica Sasanishiki wild-type (WT) genetic background. Photoperiod sensitivity, the relationship between days to heading (DTH), and yield components of mutants were investigated. We found that the yield increases with increases in DTH, but eventually plateaus at maximum and then began to decrease, whereas the biomass continued to increase. The mutants exerted distinctly different effects on DTH and yield components. The convergent double mutants had severe yield reduction compared with single mutants, even with a DTH that was similar to that of single mutants. We also found that an elite mutant of se14 achieved a yield equal to that of the WT, but with heading occurring 10 days earlier. A sequence analysis of 72 cultivars collected from the japonica cultivated zone shows that elite se14 mutants have not been applied to rice breeding. Our study demonstrates the effect of heading time genes on reproductive transition and yield components under an identical genetic background. These results may provide new insights into rice breeding using heading time mutants.





Similar content being viewed by others
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
References
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 15:819–823
Dai X, Ding Y, Tan L, Fu Y, Liu F, Zhu Z, Sun X, Gu P, Cai H, Sun C (2012) LHD1, an allele of DTH8/Ghd8, controls late heading date in common wild rice (Oryza rufipogon). J Integr Plant Biol 54:790–799
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18:926–936
Doyle J (1991) DNA protocols for plants. In: Hewitt GM, Johnston AWB, Young JPW (eds) Molecular techniques in taxonomy. NATO ASI series (Series H: Cell Biology), vol 57. Springer, Berlin, Heidelberg
Endo-Higashi N, Izawa T (2011) Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice. Plant Cell Physiol 52:1083–1094
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23:1229–1232
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832
Ishikawa R, Aoki M, Kurotani K, Yokoi S, Shinomura T, Takano M, Shimamoto K (2011) Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Genet Genom 285:461–470
Khush GS (2001) Green revolution: the way forward. Nat Rev Genet 2:815–822
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43:1096–1105
Komiya R, Ikegami A, Tamaki S et al (2003) Hd3a and RFT1 are essential for flowering in rice. Development 135:767–774
Komiya R, Yokoi S, Shimamoto K (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136:3443–3450
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li T, Chen X, Zhong X, Zhao Y, Liu X, Zhou S, Cheng S, Zhou D-X (2013) Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25:4725–4736
Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, Lin Q, Luo W, Wu G, Li H (2016) Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci 7:377
Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J (2017) A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170:114–126
Li X, Wu L, Wang J, Sun J, Xia X, Geng X, Wang X, Xu Z, Xu Q (2018) Genome sequencing of rice subspecies and genetic analysis of recombinant lines reveals regional yield- and quality-associated loci. BMC Biol 16:102
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Nishimura A, Aichi I, Matsuoka M (2006) A protocol for Agrobacterium-mediated transformation in rice. Nat Protoc 1:2796
Park SJ, Jiang K, Tal L, Yichie Y, Gar O, Zamir D, Eshed Y, Lippman ZB (2014) Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat Genet 46:1337–1342
Saito H, Okumoto Y, Yoshitake Y, Inoue H, Yuan Q, Teraishi M, Tsukiyama T, Nishida H, Tanisaka T (2011) Complete loss of photoperiodic response in the rice mutant line X61 is caused by deficiency of phytochrome chromophore biosynthesis gene. Theor Appl Genet 122:109–118
Saito H, Ogiso-Tanaka E, Okumoto Y, Yoshitake Y, Izumi H, Yokoo T, Matsubara K, Hori K, Yano M, Inoue H, Tanisaka T (2012) Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol 53:717–728
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Sun Q, Zhou D-X (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105:13679–13684
Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K (2009) Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc Natl Acad Sci 106:4555–4560
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033
Tsukada Y, Fang J, Erdjumentbromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811–816
Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153:1747–1758
Xu Q, Saito H, Hirose I, Katsura K, Yoshitake Y, Yokoo T, Tsukiyama T, Teraishi M, Tanisaka T, Okumoto Y (2014) The effects of the photoperiod-insensitive alleles, se13, hd1 and ghd7, on yield components in rice. Mol Breed 33:813–819
Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40:761–767
Yan W, Wang P, Chen H, Zhou H, Li Q, Wang C, Ding Z, Zhang YS, Yu SB, Xing YZ, Zhang QF (2011) A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant 4:319–330
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2484
Yokoo T, Saito H, Yoshitake Y, Xu Q, Asami T, Tsukiyama T, Teraishi M, Okumoto Y, Tanisaka T (2014) Se14, encoding a JmjC domain-containing protein, plays key roles in long-day suppression of rice flowering through the demethylation of H3K4me3 of RFT1. PLoS ONE 9:e96064
Yoshitake Y, Yokoo T, Saito H, Tsukiyama T, Quan X, Zikihara K, Katsura H, Tokutomi S, Aboshi T, Mori N (2015) The effects of phytochrome-mediated light signals on the developmental acquisition of photoperiod sensitivity in rice. Sci Rep 5:7709
Yuan Q, Saito H, Okumoto Y, Inoue H, Nishida H, Tsukiyama T, Teraishi M, Tanisaka T (2009) Identification of a novel gene ef7 conferring an extremely long basic vegetative growth phase in rice. Theor Appl Genet 119:675–684
Zhang J, Zhou X, Yan W, Zhang Z, Lu L, Han Z, Zhao H, Liu H, Song P, Hu Y (2015) Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol 208:1056–1066
Zhou X, Jiang G, Yang L, Qiu L, He P, Nong C, Wang Y, He Y, Xing Y (2018) Gene diagnosis and targeted breeding for blast-resistant Kongyu 131 without changing regional adaptability. J Genet Genom 45:539–547
Funding
The National Natural Science Foundation of China (31501284) supported this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Albrecht E. Melchinger.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cui, Y., Zhu, M., Xu, Z. et al. Assessment of the effect of ten heading time genes on reproductive transition and yield components in rice using a CRISPR/Cas9 system. Theor Appl Genet 132, 1887–1896 (2019). https://doi.org/10.1007/s00122-019-03324-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03324-1