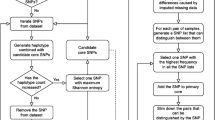
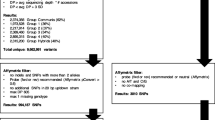
Abstract
Key message
NGS-assisted super pooling emerging as powerful tool to accelerate gene mapping and haplotype association analysis within target region uncovering specific linkage SNPs or alleles for marker-assisted gene pyramiding.
Abstract
Conventional gene mapping methods to identify genes associated with important agronomic traits require significant amounts of financial support and time. Here, a single nucleotide polymorphism (SNP)-based mapping approach, RNA-Seq and SNP array assisted super pooling analysis, was used for rapid mining of a candidate genomic region for stripe rust resistance gene Yr26 that has been widely used in wheat breeding programs in China. Large DNA and RNA super-pools were genotyped by Wheat SNP Array and sequenced by Illumina HiSeq, respectively. Hundreds of thousands of SNPs were identified and then filtered by multiple filtering criteria. Among selected SNPs, over 900 were found within an overlapping interval of less than 30 Mb as the Yr26 candidate genomic region in the centromeric region of chromosome arm 1BL. The 235 chromosome-specific SNPs were converted into KASP assays to validate the Yr26 interval in different genetic populations. Using a high-resolution mapping population (> 30,000 gametes), we confined Yr26 to a 0.003-cM interval. The Yr26 target region was anchored to the common wheat IWGSC RefSeq v1.0 and wild emmer WEWSeq v.1.0 sequences, from which 488 and 454 kb fragments were obtained. Several candidate genes were identified in the target genomic region, but there was no typical resistance gene in either genome region. Haplotype analysis identified specific SNPs linked to Yr26 and developed robust and breeder-friendly KASP markers. This integration strategy can be applied to accelerate generating many markers closely linked to target genes/QTL for a trait of interest in wheat and other polyploid species.






Similar content being viewed by others
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H et al (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178
Allen AM, Barker GLA, Berry ST, Coghill JA, Gwilliam R et al (2011) Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol J 9:1086–1099
Allen AM, Barker GLA, Wilkinson P, Burridge A, Winfield M et al (2013) Discovery and development of exome-based, co-dominant single nucleotide polymorphism markers in hexaploid wheat (Triticum aestivum L.). Plant Biotechnol J 11:279–295
Avni R, Nave M, Barad O, Baruch K, Twardziok SO et al (2017) Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357:93–97
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Brown JK (2015) Durable resistance of crops to disease: a Darwinian perspective. Annu Rev Phytopathol 53:513–539
Brueggeman R, Rostoks N, Kudrna D, Kilian A, Han F et al (2002) The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc Natl Acad Sci USA 99:9328–9333
Cao A, Xing L, Wang X, Yang X, Wang W et al (2011) Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc Natl Acad Sci USA 108:7727–7732
Chapman JA, Mascher M, Buluc AN, Barry K, Georganas E et al (2015) A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biol 16:26
Chen XM (2005) Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can J Plant Pathol 27:314–337
Chen X (2013) High-temperature adult-plant resistance, key for sustainable control of stripe rust. Am J Plant Sci 04:608–627
Chen XM (2014) Integration of cultivar resistance and fungicide application for control of wheat stripe rust. Can J Plant Pathol 36:311–326
Chen WQ, Wu LR, Liu TG, Xu SC, Jin SL et al (2009) Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis 93:1093–1101
Cheng P, Xu LS, Wang MN, See DR, Chen XM (2014) Molecular mapping of genes Yr64 and Yr65 for stripe rust resistance in hexaploid derivatives of durum wheat accessions PI 331260 and PI 480016. Theor Appl Genet 127:2267–2277
Clavijo BJ, Venturini L, Schudoma C, Accinelli GG, Kaithakottil G et al (2017) An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res 27:885–896
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ellis JG, Lagudah ES, Spielmeyer W, Dodds PN (2014) The past, present and future of breeding rust resistant wheat. Front Plant Sci 5:1–13
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L et al (2009) A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323:1357–1360
Garcia V, Bres C, Just D, Fernandez L, Tai FW et al (2016) Rapid identification of causal mutations in tomato EMS populations via mapping-by-sequencing. Nat Protoc 11:2401–2418
Giovannoni JJ, Wing RA, Ganal MW, Tanksley SD (1991) Isolation of molecular markers from specific chromosomal intervals using DNA pools from existing mapping populations. Nucleic Acids Res 19:6553–6568
Han D, Wang Q, Zhang L, Wei G, Zeng Q et al (2010) Evaluation of resistance of current wheat cultivars to stripe rust in Northwest China, North China and the Middle and Lower Reaches of Changjiang River epidemic area. Sci Agric Sin 43:2889–2896
Han DJ, Wang QL, Chen XM, Zeng QD, Wu JH et al (2015) Emerging Yr26-virulent races of Puccinia striiformis f. sp. tritici are threatening wheat production in the Sichuan Basin, China. Plant Dis 99:754–760
Hovmøller MS, Walter S, Justesen AF (2010) Escalating threat of wheat rusts. Science 329:369
Huang Q, Li X, Chen WQ, Xiang ZP, Zhong SF et al (2014) Genetic mapping of a putative Thinopyrum intermedium-derived stripe rust resistance gene on wheat chromosome 1B. Theor Appl Genet 127:843–853
Jia J, Zhao S, Kong X, Li Y, Zhao G et al (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496:91–95
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Krattinger SG, Keller B (2016) Molecular genetics and evolution of disease resistance in cereals. New Phytol 212:320–332
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J et al (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363
Li S, Chou HH (2004) LUCY2: an interactive DNA sequence quality trimming and vector removal tool. Bioinformatics 20:2865–2866
Li ZQ, Zeng SM (eds) (2002) Wheat rust in China. China Agriculture Press, Beijing
Li GQ, Li ZF, Yang WY, Zhang Y, He ZH et al (2006) Molecular mapping of stripe rust resistance gene YrCH42 in Chinese wheat cultivar Chuanmai 42 and its allelism with Yr24 and Yr26. Theor Appl Genet 112:1434–1440
Li Q, Ma D, Li Q, Fan Y, Shen X et al (2016) Genetic analysis and molecular mapping of a stripe rust resistance gene in Chinese wheat differential Guinong 22. J Phytopathol 164:476–484
Li B, Xu Q, Yang Y, Wang Q, Zeng Q et al (2017) Stripe rust resistance and genes in Chongqing wheat cultivars and lines. Sci Agric Sin 50:413–425
Lin F, Chen XM (2007) Genetics and molecular mapping of genes for race-specific all-stage resistance and non-race-specific high-temperature adult-plant resistance to stripe rust in spring wheat cultivar Alpowa. Theor Appl Genet 114:1277–1287
Lin F, Chen XM (2008) Molecular mapping of genes for race-specific overall resistance to stripe rust in wheat cultivar Express. Theor Appl Genet 116:797–806
Line RF, Qayoum A (1992) Virulence, aggressiveness, evolution, and distribution of races of Puccinia striiformis (the cause of stripe rust of wheat) in North America 1968–1987. US Department of Agriculture Technical Bulletin No. 1788, p 74
Ling HQ, Zhao S, Liu D, Wang J, Sun H et al (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496:87–90
Liu S, Yeh C, Tang HM, Nettleton D, Schnable PS (2012) Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS ONE 7:e36406
Liu W, Frick M, Huel R, Nykiforuk CL, Wang X et al (2014) The stripe rust resistance gene Yr10 encodes an evolutionary-conserved and unique CC-NBS-LRR sequence in wheat. Mol Plant 7:1740–1755
Liu B, Liu T, Zhang Z, Jia Q, Wang B et al (2017) Discovery and pathogenicity of CYR34, a new race of Puccinia striiformis f. sp. tritici in China. Acta Phytopathol Sin. https://doi.org/10.13926/j.cnki.apps.000071
Ma J, Zhou R, Dong Y, Wang L, Wang X et al (2001) Molecular mapping and detection of the yellow rust resistance gene Yr26 in wheat transferred from Triticum turgidum L. using microsatellite markers. Euphytica 120:219–226
McIntosh RA, Lagudah ES (2000) Cytogenetical studies in wheat. XVIII. Gene Yr24 for resistance to stripe rust. Plant Breeding 119:81–83
McIntosh RA, Dubcovsky J, Rogers J, Morris C, Appels R et al. (2016) Catalogue of gene symbols for wheat: 2016 Supplement. https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2015.pdf. Accessed 20 Sept 2017
McIntosh RA, Dubcovsky J, Rogers J, Morris C, Appels R et al. (2017) Catalogue of gene symbols for wheat: 2017 Supplement. https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2017.pdf. Accessed 20 Sept 2017
McIntosh RA, Mu J, Han D, Kang Z (2018) Wheat stripe rust resistance gene Yr24/Yr26: a retrospective review. Crop J. https://doi.org/10.1016/j.cj.2018.02.001
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Moore JW, Herrera-Foessel S, Lan C, Schnippenkoetter W, Ayliffe M et al (2015) A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat Genet 47:1494–1498
Nagy ED, Eder C, Molnár-Láng M, Lelley T (2003) Genetic mapping of sequence-specific PCR-based markers on the short arm of the 1BL.1RS wheat-rye translocation. Euphytica 132:243–249
Peng JH, Fahima T, Der Röder MS, Li YC, Dahan A et al (1999) Microsatellite tagging of the stripe-rust resistance gene YrH52 derived from wild emmer wheat, Triticum dicoccoides, and suggestive negative crossover interference on chromosome 1B. Theor Appl Genet 98:862–872
Periyannan S, Milne RJ, Figueroa M, Lagudah ES, Dodds PN (2017) An overview of genetic rust resistance: from broad to specific mechanisms. PLoS Pathog 13:e1006380
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ramirez-Gonzalez RH, Segovia V, Bird N, Fenwick P, Holdgate S et al (2015a) RNA-Seq bulked segregant analysis enables the identification of high-resolution genetic markers for breeding in hexaploid wheat. Plant Biotechnol J 13:613–624
Ramirez-Gonzalez RH, Uauy C, Caccamo M (2015b) PolyMarker: a fast polyploid primer design pipeline. Bioinformatics 31:2038–2039
Rasheed A, Hao Y, Xia X, Khan A, Xu Y et al (2017) Crop breeding chips and genotyping platforms: progress, challenges, and perspectives. Mol plant 10:1047–1064
Philippe R, Paux E, Bertin I, Sourdille P, Choulet F, Laugier C, Simková H, Safář J, Bellec A, Vautrin S, Frenkel Z, Cattonaro F, Magni F, Scalabrin S, Martis MM, Mayer KF, Korol A, Bergès H, Doležel J, Feuillet C (2013) A high density physical map of chromosome 1BL supports evolutionary studies, map-based cloning and sequencing in wheat. Genome Biol 14(6):R64
Schlötterer C, Tobler R, Kofler R, Nolte V (2014) Sequencing pools of individuals—mining genome-wide polymorphism data without big funding. Nat Rev Genet 15:749–763
Singh VK, Khan AW, Saxena RK, Kumar V, Kale SM et al (2016) Next-generation sequencing for identification of candidate genes for Fusarium wilt and sterility mosaic disease in pigeonpea (Cajanus cajan). Plant Biotechnol J 14:1183–1194
Song WN, Ko L, Henry RJ (1994) Polymorphisms in the α-amy1 gene of wild and cultivated barley revealed by the polymerase chain reaction. Theor Appl Genet 89:509–513
St. Clair DA (2010) Quantitative disease resistance and quantitative resistance loci in breeding. Annu Rev Phytopathol 48:247–268
Steuernagel B, Periyannan SK, Hernández-Pinzón I, Witek K, Rouse MN et al (2016) Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol 34:652–655
Stubbs RW (1985) Stripe rust. In: Roelfs AP, Bushnell WR (eds) The cereal rusts, vol II. Academic Press, New York, pp 61–101
Sun GL, Fahima T, Korol AB, Turpeinen T, Grama A et al (1997) Identification of molecular markers linked to the Yr15 stripe rust resistance gene of wheat originated in wild emmer wheat, Triticum dicoccoides. Theor Appl Genet 95:622–628
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S et al (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183
Tamura K, Peterson D, Peterson N, Stecher G, Nei M et al (2011) MEGA5: molecular revolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thind AK, Wicker T, Simkova H, Fossati D, Moullet O et al (2017) Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nat Biotechnol 35:793–796
Trick M, Adamski N, Mugford SG, Jiang C, Febrer M et al (2012) Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol 12:14
Uauy C (2017) Wheat genomics comes of age. Curr Opin Plant Biol 36:142–148
Uauy C, Wulff BBH, Dubcovsky J (2017) Combining traditional mutagenesis with new high-throughput sequencing and genome editing to reveal hidden variation in polyploid wheat. Annu Rev Genet 51:435–454
United Nations Department Of Economic And Social Affairs PD (2015) World population prospects: the 2015 revision. Working paper no. ESA/P/WP.241. https://esa.un.org/unpd/wpp/. Accessed 27 Feb 2017 (WWW document)
Uricaru R, Rizk G, Lacroix V, Quillery E, Plantard O et al (2015) Reference-free detection of isolated SNPs. Nucleic Acids Res 43:e11
Van Ooijen JW (2006) JoinMap4, software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
van Poecke RMP, Maccaferri M, Tang J, Truong HT, Janssen A et al (2013) Sequence-based SNP genotyping in durum wheat. Plant Biotechnol J 11:809–817
Varshney RK, Terauchi R, McCouch SR (2014) Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol 12:e1001883
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang L, Ma J, Zhou R, Wang X, Jia J (2002) Molecular tagging of the yellow rust resistance gene Yr10 in common wheat, PI178383 (Triticum aestivum L.). Euphytica 124:71–73
Wang C, Zhang Y, Han D, Kang Z, Li G et al (2008) SSR and STS markers for wheat stripe rust resistance gene Yr26. Euphytica 159:359–366
Wang S, Wong D, Forrest K, Allen A, Chao S et al (2014) Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wang Y, Xie J, Zhang H, Guo B, Ning S et al (2017) Mapping stripe rust resistance gene YrZH22 in Chinese wheat cultivar Zhoumai 22 by bulked segregant RNA-Seq (BSR-Seq) and comparative genomics analyses. Theor Appl Genet 130:2191–2201
Wellings CR (2011) Global status of stripe rust: a review of historical and current threats. Euphtica 179:129–141
William M, Singh RP, Huerta-Espino J, Islas SO, Hoisington D (2003) Molecular marker mapping of leaf rust resistance gene Lr46 and its association with stripe rust resistance gene Yr29 in wheat. Phytopathology 93:153–159
Wu TD, Nacu S (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873–881
Wu JH, Wang QL, Chen XM, Wang MJ, Mu JM et al (2016) Stripe rust resistance in wheat breeding lines developed for central Shaanxi, an overwintering region for Puccinia striiformis f. sp. tritici in China. Can J Plant Pathol 38:317–324
Wu J, Liu S, Wang Q, Zeng Q, Mu J et al (2018a) Rapid identification of an adult plant stripe rust resistance gene in hexaploid wheat by high-throughput SNP array genotyping of pooled extremes. Theor Appl Genet 131:43–58
Wu J, Wang Q, Xu L, Chen X, Li B et al (2018b) Combining SNP genotyping array with bulked segregant analysis to map a gene controlling adult-plant resistance to stripe rust in wheat line 03031-1-5 H62. Phytopathology 108:103–113
Xu Y, Li P, Zou C, Lu Y, Xie C et al (2017) Enhancing genetic gain in the era of molecular breeding. J Exp Bot 68:2641–2666
Zeng Q, Han D, Wang Q, Yuan F, Wu J et al (2014) Stripe rust resistance and genes in Chinese wheat cultivars and breeding lines. Euphytica 196:271–284
Zhang X, Han D, Zeng Q, Duan Y, Yuan F et al (2013) Fine mapping of wheat stripe rust resistance gene Yr26 based on collinearity of wheat with Brachypodium distachyon and rice. PLoS ONE 8:e57885
Zhang H, Zhang L, Wang C, Wang Y, Zhou X et al (2016) Molecular mapping and marker development for the Triticum dicoccoides-derived stripe rust resistance gene YrSM139-1B in bread wheat cv. Shaanmai 139. Theor Appl Genet 129:369–376
Zou C, Wang P, Xu Y (2016) Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol J 14:1941–1955
Acknowledgements
The authors are grateful to Prof. R.A. McIntosh, Plant Breeding Institute, University of Sydney, for critical review of this manuscript; Prof. Peidu Chen and Prof. Aizhong Cao, Cytogenetics Institute, Nanjing Agricultural University, for providing Yr26 germplasms and genetic populations. This study was financially supported by International S&T Cooperation Program of China (2015DFG32340), National Natural Science Foundation of China (31371924), the National Key Research and Development Program of China (Grant no. 2016YFE0108600), the earmarked funds for Modern Agro-industry Technology Research System (No. CARS-3-1-11) and National Natural Science Foundation for Young Scientists of China (Grant 31701421).
Author information
Authors and Affiliations
Contributions
JHW designed and conducted the experiments, analyzed the data, and wrote the manuscript. QDZ analyzed the data, prepared the figures for the manuscript and contributed to writing the RNA-Seq sections; QLW participated in creating the genetic populations and analyzed the SNP array data. SJL, JMM and SH participated in greenhouse and field experiments and contributed to the genotyping experiment. SZY assisted in analyzing the data and prepared the figures for the manuscript. HS and AD analyzed the data with the wild emmer genome. LLH participated in revising the manuscript. DJH and ZSK conceived and directed the project and revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Ethical standard
I declare on behalf of my co-authors that the work described is original, previously unpublished research, and not under consideration for publication elsewhere. The experiments in this study comply with the current laws of China.
Additional information
Communicated by Thomas Miedaner.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, J., Zeng, Q., Wang, Q. et al. SNP-based pool genotyping and haplotype analysis accelerate fine-mapping of the wheat genomic region containing stripe rust resistance gene Yr26. Theor Appl Genet 131, 1481–1496 (2018). https://doi.org/10.1007/s00122-018-3092-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3092-8