Abstract
Key message
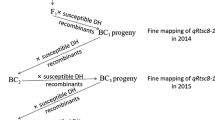
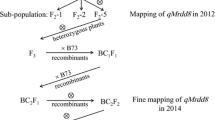
Msv1 , the major QTL for MSV resistance was delimited to an interval of 0.87 cM on chromosome 1 at 87 Mb and production markers with high prediction accuracy were developed.
Abstract
Maize streak virus (MSV) disease is a devastating disease in the Sub-Saharan Africa (SSA), which causes significant yield loss in maize. Resistance to MSV has previously been mapped to a major QTL (Msv1) on chromosome 1 that is germplasm and environment independent and to several minor loci elsewhere in the genome. In this study, Msv1 was fine-mapped through QTL isogenic recombinant strategy using a large F 2 population of CML206 × CML312 to an interval of 0.87 cM on chromosome 1. Genome-wide association study was conducted in the DTMA (Drought Tolerant Maize for Africa)-Association mapping panel with 278 tropical/sub-tropical breeding lines from CIMMYT using the high-density genotyping-by-sequencing (GBS) markers. This study identified 19 SNPs in the region between 82 and 93 Mb on chromosome 1(B73 RefGen_V2) at a P < 1.00E-04, which coincided with the fine-mapped region of Msv1. Haplotype trend regression identified a haplotype block significantly associated with response to MSV. Three SNPs in this haplotype block at 87 Mb on chromosome 1 had an accuracy of 0.94 in predicting the disease reaction in a collection of breeding lines with known responses to MSV infection. In two biparental populations, selection for resistant Msv1 haplotype demonstrated a reduction of 1.03–1.39 units on a rating scale of 1–5, compared to the susceptible haplotype. High-throughput KASP assays have been developed for these three SNPs to enable routine marker screening in the breeding pipeline for MSV resistance.





Similar content being viewed by others
References
Abalo G, Tongoona P, Derera J, Edema R (2009) A comparative analysis of conventional and marker-assisted selection methods in breeding maize streak virus resistance in maize. Crop Sci 49:509–520
Bent AF (1996) Plant disease resistance genes: function meets structure. Plant Cell 8:1757–1771
Bishop DT, Williamson JA (1990) The power of identity-by-state methods for linkage analysis. Am J Hum Genet 46:254–265
Cairns JE, Crossa J, Zaidi PH et al (2013) Identification of drought, heat, and combined drought and heat tolerant donors in maize. Crop Sci 53:1335–1346
Ching A, Caldwell KS, Jung M et al (2002) SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet 3:19
CIMMYT (2001) Laboratory protocols: CIMMYT applied molecular genetics laboratory protocols. CIMMYT, Mexico
Elshire RJ, Glaubitz JC, Sun Q et al (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6(5):e19379
Ersoz E, Yu J, Buckler E (2009) Applications of linkage disequilibrium and association mapping in maize. In: Kirz A, Larkins B (eds) Molecular genetic approaches to maize improvement. Springer, Berlin, pp 173–195
Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927
Gabriel SB, Schaffner SF, Nguyen H et al (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229
Gao S, Martinez C, Skinner DJ et al (2008) Development of a seed DNA-based genotyping system for marker-assisted selection in maize. Mol Breed 22:477–494
Guthrie EJ (1978) Measurement of yield losses caused by maize streak disease. Plant Dis Rep 62:839–840
Hill WG, Weir BS (1988) Variances and covariances of squared linkage disequilibria in finite populations. Theor Popul Biol 33:54–78
Johnson GCL, Esposito L, Barratt BJ et al (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29:233–237
Kim SK, Efron Y, Khadr F, Fajemisin J, Lee MH (1987) Registration of 16 maize streak resistant tropical maize parental inbred lines. Crop Sci 27:824–825
Kim SK, Efron Y, Fajemisin JM, Buddenhagen IW (1989) Mode of gene action for resistance in maize to maize streak virus. Crop Sci 29:890–894
Kyetere DT, Ming R, McMullen MD et al (1999) Genetic analysis of tolerance to maize streak virus in maize. Genome 42:20–26
Lagat M, Danson M, Kimani M, Kuria A (2008) Quantitative trait loci for resistance to maize streak virus in maize genotypes used in hybrid development. Afric J Biotech 7:2573–2577
Martin DP, Shepherd DN (2009) The epidemiology, economic impact and control of maize streak disease. Food Sec 1:305–315
Martin DP, Willment JA, Billharz R et al (2001) Sequence diversity and virulence in Zea mays of Maize streak virus isolates. Virology 288:247–255
Myles S, Peiffer J, Brown P et al (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21:2194–2202
Nordborg M, Weigel D (2008) Next-generation genetics in plants. Nature 456:720–723
Peleman JD, Wye C, Zethof J et al (2005) Quantitative trait locus (QTL) isogenic recombinant analysis: a method for high resolution mapping of QTL within a single population. Genetics 171:1341–1352
Pernet A, Hoisington D, Franco J et al (1999a) Genetic mapping of maize streak virus resistance from the Mascarene source. I. Resistance in line D211 and stability against different virus clones. Theor Appl Genet 99:524–539
Pernet A, Hoisington D, Dintinger J et al (1999b) Genetic mapping of maize streak virus resistance from the Mascarene source. II. Resistance in line CIRAD390 and stability across germplasm. Theor Appl Genet 99:540–553
Prasanna BM, Chaikam V, Mahuku G (2012) Doubled haploid (DH) technology in maize breeding: an overview. In: Prasanna BM, Chaikam V, Mahuku G (eds) Doubled haploid technology in maize breeding: theory and practice. CIMMYT, Mexico, pp 1–8
Prasanna BM, Babu R, Nair S et al (2014) Molecular marker-assisted breeding for tropical maize improvement. In: Wusurika R, Bohn M, Lai J, Kole C (eds) Genetics, genomics and breeding of maize. CRC Press, London, pp 89–119
Pratt RC, Gordon SG (2006) Breeding for resistance to maize foliar pathogens. Plant Breed Rev 27:119–174
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/
Rafalski A (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 5:94–100
Rafalski JA (2010) Association genetics in crop improvement. Curr Opin Plant Biol 13:174–180
Remington DL, Thornsberry JM, Matsuoka Y et al (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Nat Acad Sci USA 98:11479–11484
Rodier A, Assie J, Marchand JL, Herve Y (1995) Breeding maize lines for complete and partial resistance to maize streak virus (MSV). Euphytica 81:57–70
Romay MC, Millard MJ, Glaubitz JC et al (2013) Comprehensive genotyping of the USA national maize inbred seeds bank. Genome Biol 14:R55
Semagn K, Beyene Y, Makumbi D et al (2012) Quality control genotyping for assessment of genetic identity and purity in diverse tropical maize inbred lines. Theor Appl Genet 125:1487–1501
Shepherd DN, Mangwende T, Martin DP et al (2007) Maize streak virus-resistant transgenic maize: a first for Africa. Plant Biotech J 5:759–767
Shepherd DN, Martin DP, Van der walt E et al (2010) Maize streak virus: an old and complex ‘emerging’ pathogen. Mol Plant Path 11:1–12
Soto PE, Buddenhagen IW, Asnani VL (1982) Development of streak virus-resistant maize populations through improved challenge and selection methods. Ann Appl Biol 100:539–546
Storey HH, Howland AK (1967) Inheritance of resistance in maize to the virus of streak disease in East Africa. Ann Appl Biol 59:429–436
Stuber CW, Lincoln SF, Wolff DW (1987) Molecular markers facilitated investigations of quantitative trait loci in maize. II. Factors influencing yield and its component traits. Crop Sci 27:639–648
Tang CY, Bjarnason MJ (1993) Two approaches for the development of maize germplasm resistant to maize streak virus. Maydica 38:301–307
Tenaillon MI, Sawkins MC, Long AD et al (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci USA 98:9161–9166
Wambugu F, Wafula J (2000) Advances in maize streak virus disease research in Eastern and Southern Africa. Workshop Report, 15–17 September 1999, KARI and ISAAA AfriCenter, Nairobi, Kenya. ISAAA Briefs No. 16. ISAAA, Ithaca, NY, p 43
Welz HG, Schechert A, Pernet A et al (1998) A gene for resistance to maize streak virus in the African CIMMYT maize inbred CML202. Mol Breed 4:147–154
Yan J, Shah T, Warburton M et al (2009) Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS One 04:e8451
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178:539–551
Zeng LR, Vega-Sanchez ME, Zhu T, Wang GL (2006) Ubiquitinization-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res 16:413–426
Zhu C, Gore M, Buckler E, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Acknowledgments
The authors gratefully acknowledge the financial support received from the Bill and Melinda Gates Foundation (BMGF) as part of the project, “Drought Tolerant Maize for Africa (DTMA)”. We thank CGIAR Research Program (CRP) on MAIZE for co-sponsoring this research work. The biparental populations and disease data from Kevin Pixley, CIMMYT and Jean-Marcel Ribaut, GCP, used in initial QTL mapping and validation study are thankfully acknowledged. Assistance for data analysis provided by Jyothsna Tejomurthula is appreciated. The authors would also like to thank the technical assistance from Carlos Martinez, Alberto Vergara and Jose Simon Marias of CIMMYT in carrying out this research work. The authors also gratefully acknowledge the two reviewers for their valuable comments that could improve the value of this manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare no ethical standards have been violated in the course of the study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Frisch.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nair, S.K., Babu, R., Magorokosho, C. et al. Fine mapping of Msv1, a major QTL for resistance to Maize Streak Virus leads to development of production markers for breeding pipelines. Theor Appl Genet 128, 1839–1854 (2015). https://doi.org/10.1007/s00122-015-2551-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2551-8