Abstract
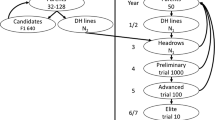
QTL detection is a good way to assess the genetic basis of quantitative traits such as the plant response to its environment, but requires large mapping populations. Experimental constraints, however, may require a restriction of the population size, risking a decrease in the quality level of QTL mapping. The purpose of this paper was to test if an advanced backcross population sample chosen by MapPop 1.0 could limit the effect of size restriction and improve the QTL detection when compared to random samples. We used the genotypic and phenotypic data obtained for 280 genotypes, considered as the reference population. The “MapPop sample” of 100 genotypes was first compared to the reference population, and genetic maps, genotypic and phenotypic data and QTL results were analysed. Despite the increase in donor allele frequency in the MapPop sample, this did not lead to an increase of the genetic map length or a biased phenotypic distribution. Three QTL among the 10 QTL found in the reference population were also detected in the MapPop sample. Next, the MapPop sample results were compared to those from 500 random samples of the same size. The main conclusion was that the MapPop software avoided the selection of biased samples and the detection of false QTL and appears particularly interesting to select a sample from an unbalanced population.






Similar content being viewed by others
References
Asins MJ (2002) Present and future of quantitative trait locus analysis in plant breeding. Plant Breed 121:281–291
Bernacchi D, Beck-Bunn T, Emmatty D, Eshed Y, Inai S, Lopez J, Petiard V, Sayama H, Uhlig J, Zamir D, Tanksley S (1998a) Advanced backcross QTL analysis of tomato. II. Evaluation of near-isogenic lines carrying single-donor introgressions for desirable wild QTL-alleles derived from Lycopersicon hirsutum and L. pimpinellifolium. Theor Appl Genet 97:170–180
Bernacchi D, Beck-Bunn T, Eshed Y, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley S (1998b) Advanced backcross QTL analysis in tomato. I. Identification of QTLs for traits of agronomic importance from Lycopersicon hirsutum. Theor Appl Genet 97:381–397
Bernardo R (2004) What proportion of declared QTL in plants are false? Theor Appl Genet 109:419–24
Charcosset A, Gallais A (1996) Estimation of the contribution of quantitative trait loci (QTL) to the variance of a quantitative trait by means of genetic markers. Theor Appl Genet 93:1193–1201
Chardon F, Virlon B, Moreau L, Falque M, Joets J, Decousset L, Murigneux A, Charcosset A (2004) Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics 168:2169–2185
Charmet G (2000) Power and accuracy of QTL detection: simulation studies of one-QTL models. Agronomie 20:309–323
Doerge R (2002) Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet 3:43–52
Doganlar S, Frary A, Ku H, Tanksley S (2002) Mapping quantitative trait loci in inbred backcross lines of Lycopersicon pimpinellifolium (LA1589). Genome 45:1189–1202
Fracheboud Y, Jompuk C, Ribaut J, Stamp P, Leipner J (2004) Genetic analysis of cold-tolerance of photosynthesis in maize. Plant Mol Biol 56:241–253
Gallais A, Rives M (1993) Detection, number and effects of QTLs for a complex character. Agronomie 13:723–738
Haley C, Knott S (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69:315–324
Hyne V, Kearsey M, Pike D, Snape J (1995) QTL analysis: unreliability and bias in estimation procedures. Mol Breed 1:273–282
Jannink JL (2005) Selective phenotyping to accurately map quantitative trait loci. Crop Sci 45:901–908
Jin C, Lan H, Attie AD, Churchill GA, Bulutuglo D, Yandell BY (2004) Selective phenotyping for increased efficiency in genetic mapping study. Genetics 168:2285–2293
Lander E, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genomics 121:185–199
Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lewis RS, Goodman MM (2003) Incorporation of tropical maize germplasm into inbred lines derived from temperate x temperate-adapted tropical line crosses: agronomic and molecular assessment. Theor Appl Genet 107:798–805
Lincoln S, Daly M, Lander E (1992) Constructing genetic maps with MAPMAKER/EXP3.0, 3rd edn. Whitehead Institute Technical Reports
Moncada P, Martínez CP, Borrero J, Chatel M, Guimaraes HGJrE, Tohme J, McCouch SR (2001) Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet 102:41–52
Paterson A, Saranga Y, Menz M, Jiang C, Wright R (2003) QTL analysis of genotype × environment interactions affecting cotton fiber quality. Theor Appl Genet 106:384–396
Piepho H (2000) Optimal marker density for interval mapping in a backcross population. Heredity 84:437–440
Pillen K, Zacharias A, Leon J (2003) Advanced backcross QTL analysis in barley (Hordeum vulgare L.). Theor Appl Genet 107:340–352
Pillen K, Zacharias A, Leon J (2004) Comparative AB-QTL analysis in barley using a single exotic donor of Hordeum vulgare ssp spontaneum. Theor Appl Genet 108:1591–1601
SAS institute Inc (1991) SAS user’s guide: release 8.1 eds SAS Institute, Cary, North Carolina
Servin B, Dillmann C, Decoux G, Hospital F (2002) MDM: a program to compute fully informative genotype frequencies in complex breeding schemes. J Hered 93:227–228
Soller M, Brody T, Genizi A (1976) On the power of experimental designs for the detection of linkage between marker loci and quantitative loci in crosses between inbred lines. Theor Appl Genet 47:35–39
Tanksley S, Nelson J (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Tanksley SD, Grandillo S, Fulton TM, Zamir D, Eshed T, Petiard V, Lopez J, Beck-Bunn T (1996) Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor Appl Genet 92:213–224
Tuberosa R, Salvi S, Sanguineti M, Maccaferri M, Giuliani S, Landi P (2003) Searching for quantitative trait loci controlling root traits in maize: a critical appraisal. Plant Soil 255:35–54
Utz H, Melchinger A, Schon C (2000) Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics 154:1839–1849
Vales M, Schon C, Capettini F, Chen X, Corey A, Mather D, Mundt C, Richardson K, Sandoval-Islas J, Utz H, Hayes P (2005) Effect of population size on the estimation of QTL: a test using resistance to barley stripe rust. Theor Appl Genet 111:1260–1270
Van Ooijen J, Voorrips R (2001) Joinmap 3.0, software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Vision T, Brown D, Shmoys D, Durrett R, Tanksley S (2000) Selective mapping: a strategy for optimizing the construction of high-density linkage maps. Genetics 155:407–420
Voorrips R (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Xu Z, Zou F, Vision T (2005) Improving quantitative trait loci mapping resolution in experimental crosses by the use of genotypically selected samples. Genetics 170:401–408
Acknowledgments
The data collection was supported by Genoplante, the French consortium for plant genomics (http://www.genoplante.com). The data analysis was part of a PhD work granted by the Conseil Régional de Picardie (http://www.cr-picardie.fr)
We thank A.-L. Lainé and N. Bahrman for their technical advises for genotyping. We thank P. Barre of INRA, Lusignan, France and J.C. Nelson of Department of Plant Pathology, Kansas State University, USA for their technical and scientific help for the genetic mapping. J.C. Nelson supplies the QMap software. We thank S. Carof, X. Charrier, D. Denoue, M. Dupin, F. Lardière, and G. Leau for their contribution to the phenotyping of the population in the different locations. We thank Theo Hendricks for his valuable comments and suggestions on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.-L. Jannink.
Rights and permissions
About this article
Cite this article
Birolleau-Touchard, C., Hanocq, E., Bouchez, A. et al. The use of MapPop1.0 for choosing a QTL mapping sample from an advanced backcross population. Theor Appl Genet 114, 1019–1028 (2007). https://doi.org/10.1007/s00122-006-0495-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0495-8