Abstract
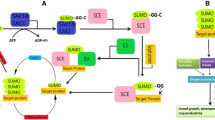
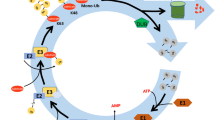
Protein post-translational modifications diversify the proteome and install new regulatory levels that are crucial for the maintenance of cellular homeostasis. Over the last decade, the ubiquitin-like modifying peptide small ubiquitin-like modifier (SUMO) has been shown to regulate various nuclear processes, including transcriptional control. In plants, the sumoylation pathway has been significantly implicated in the response to environmental stimuli, including heat, cold, drought, and salt stresses, modulation of abscisic acid and other hormones, and nutrient homeostasis. This review focuses on the emerging importance of SUMO in the abiotic stress response, summarizing the molecular implications of sumoylation and emphasizing how high-throughput approaches aimed at identifying the full set of SUMO targets will greatly enhance our understanding of the SUMO–abiotic stress association.



Similar content being viewed by others
References
Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22:159–180. doi:10.1146/annurev.cellbio.22.010605.093503
Vertegaal AC (2011) Uncovering ubiquitin and ubiquitin-like signaling networks. Chem Rev 111:7923–7940. doi:10.1021/cr200187e
Downes B, Vierstra RD (2005) Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem Soc Trans 33:393–399. doi:10.1042/BST0330393
Miura K, Hasegawa PM (2010) Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol 20:223–232. doi:10.1016/j.tcb.2010.01.007
Hay RT (2005) SUMO: a history of modification. Mol Cell 18:1–12. doi:10.1016/j.molcel.2005.03.012
Lomeli H, Vazquez M (2011) Emerging roles of the SUMO pathway in development. Cell Mol Life Sci 68:4045–4064. doi:10.1007/s00018-011-0792-5
Meulmeester E, Melchior F (2008) Cell biology: SUMO. Nature 452:709–711. doi:10.1038/452709a
Wilkinson KA, Henley JM (2010) Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428:133–145. doi:10.1042/BJ20100158
Geoffroy MC, Hay RT (2009) An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol 10:564–568. doi:10.1038/nrm2707
Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD (2010) Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA 107:16512–16517. doi:10.1073/pnas.1004181107
Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT (2009) System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2:ra24. doi:10.1126/scisignal.2000282
Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H (2008) Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 10:547–555. doi:10.1038/ncb1717
Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, Liebler DC (2004) Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol 17:1706–1715. doi:10.1021/tx049767l
Zhou W, Ryan JJ, Zhou H (2004) Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem 279:32262–32268. doi:10.1074/jbc.M404173200
Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278:6862–6872. doi:10.1074/jbc.M209694200
Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ, Hasegawa PM (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102:7760–7765. doi:10.1073/pnas.0500778102
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19:1403–1414. doi:10.1105/tpc.106.048397
Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106:5418–5423. doi:10.1073/pnas.0811088106
Miura K, Sato A, Ohta M, Furukawa J (2011) Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2. Planta 234:1191–1199. doi:10.1007/s00425-011-1476-y
Miura K, Lee J, Gong Q, Ma S, Jin JB, Yoo CY, Miura T, Sato A, Bohnert HJ, Hasegawa PM (2011) SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiol 155:1000–1012. doi:10.1104/pp.110.165191
Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19:2952–2966. doi:10.1105/tpc.106.049981
Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM (2006) SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 142:1548–1558. doi:10.1104/pp.106.088831
Cheong MS, Park HC, Hong MJ, Lee J, Choi W, Jin JB, Bohnert HJ, Lee SY, Bressan RA, Yun DJ (2009) Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol 151:1930–1942. doi:10.1104/pp.109.143719
Park BS, Song JT, Seo HS (2011) Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat Commun 2:400. doi:10.1038/ncomms1408
Chen C, Chen Y, Tang I, Liang H, Lai C, Chiou J, Yeh K (2011) Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance. Plant Physiol 156:2225–2234. doi:10.1104/pp.111.178996
Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A (2008) Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20:2894–2908. doi:10.1105/tpc.108.058669
Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007) Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145:119–134. doi:10.1104/pp.107.102285
Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, Kim D, Lee SY, Salt DE, Mengiste T, Gong Q, Ma S, Bohnert HJ, Kwak SS, Bressan RA, Hasegawa PM, Yun DJ (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49:79–90. doi:10.1111/j.1365-313X.2006.02947.x
Miura K, Lee J, Miura T, Hasegawa PM (2010) SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol 51:103–113. doi:10.1093/pcp/pcp171
van den Burg HA, Kini RK, Schuurink RC, Takken FL (2010) Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 22:1998–2016. doi:10.1105/tpc.109.070961
Park HC, Choi W, Park HJ, Cheong MS, Koo YD, Shin G, Chung WS, Kim WY, Kim MG, Bressan RA, Bohnert HJ, Lee SY, Yun DJ (2011) Identification and molecular properties of SUMO-binding proteins in Arabidopsis. Mol Cells 32:143–151. doi:10.1007/s10059-011-2297-3
Burroughs AM, Balaji S, Iyer LM, Aravind L (2007) Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct 2:18. doi:10.1186/1745-6150-2-18
Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol 280:275–286. doi:10.1006/jmbi.1998.1839
Miura K, Jin JB, Hasegawa PM (2007) Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 10:495–502. doi:10.1016/j.pbi.2007.07.002
Reed JM, Dervinis C, Morse AM, Davis JM (2010) The SUMO conjugation pathway in Populus: genomic analysis, tissue-specific and inducible SUMOylation and in vitro de-SUMOylation. Planta 232:51–59. doi:10.1007/s00425-010-1151-8
Budhiraja R, Hermkes R, Muller S, Schmidt J, Colby T, Panigrahi K, Coupland G, Bachmair A (2009) Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol 149:1529–1540. doi:10.1104/pp.108.135053
Colby T, Matthai A, Boeckelmann A, Stuible HP (2006) SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142:318–332. doi:10.1104/pp.106.085415
Castano-Miquel L, Segui J, Lois LM (2011) Distinctive properties of Arabidopsis SUMO paralogues support the in vivo predominant role of AtSUMO1/2 isoforms. Biochem J 436:581–590. doi:10.1042/BJ20101446
Gareau JR, Lima CD (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11:861–871. doi:10.1038/nrm3011
Bossis G, Melchior F (2006) SUMO: regulating the regulator. Cell Div 1:13. doi:10.1186/1747-1028-1-13
Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, Yun DJ, Bressan RA, Hasegawa PM (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 53:530–540. doi:10.1111/j.1365-313X.2007.03359.x
Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q, Yang C (2009) The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60:666–678. doi:10.1111/j.1365-313X.2009.03992.x
Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21:2284–2297. doi:10.1105/tpc.109.068072
Chosed R, Mukherjee S, Lois LM, Orth K (2006) Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem J 398:521–529. doi:10.1042/BJ20060426
Lois LM (2010) Diversity of the SUMOylation machinery in plants. Biochem Soc Trans 38:60–64. doi:10.1042/BST0380060
Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G (2003) A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15:2308–2319. doi:10.1105/tpc.015487
Park HJ, Kim WY, Park HC, Lee SY, Bohnert HJ, Yun DJ (2011) SUMO and SUMOylation in plants. Mol Cells 32:305–316. doi:10.1007/s10059-011-0122-7
Miura K, Ohta M (2010) SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J Plant Physiol 167:555–560. doi:10.1016/j.jplph.2009.11.003
Miller MJ, Vierstra RD (2011) Mass spectrometric identification of SUMO substrates provides insights into heat stress-induced SUMOylation in plants. Plant Signal Behav 6:130–133
Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ (2009) PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol Life Sci 66:3029–3041. doi:10.1007/s00018-009-0061-z
Park HC, Kim H, Koo SC, Park HJ, Cheong MS, Hong H, Baek D, Chung WS, Kim DH, Bressan RA, Lee SY, Bohnert HJ, Yun DJ (2010) Functional characterization of the SIZ/PIAS-type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice. Plant Cell Environ 33:1923–1934. doi:10.1111/j.1365-3040.2010.02195.x
Novatchkova M, Budhiraja R, Coupland G, Eisenhaber F, Bachmair A (2004) SUMO conjugation in plants. Planta 220:1–8. doi:10.1007/s00425-004-1370-y
Makhnevych T, Sydorskyy Y, Xin X, Srikumar T, Vizeacoumar FJ, Jeram SM, Li Z, Bahr S, Andrews BJ, Boone C, Raught B (2009) Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol Cell 33:124–135. doi:10.1016/j.molcel.2008.12.025
Okada S, Nagabuchi M, Takamura Y, Nakagawa T, Shinmyozu K, Nakayama J, Tanaka K (2009) Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol 50:1049–1061. doi:10.1093/pcp/pcp056
Cohen-Peer R, Schuster S, Meiri D, Breiman A, Avni A (2010) Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotholerance. Plant Mol Biol 74:33–45. doi:10.1007/s11103-010-9652-1
Gill G (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15:536–541. doi:10.1016/j.gde.2005.07.004
Lyst MJ, Stancheva I (2007) A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans 35:1389–1392. doi:10.1042/BST0351389
Garcia-Dominguez M, Reyes JC (2009) SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta 1789:451–459. doi:10.1016/j.bbagrm.2009.07.001
Garcia-Dominguez M, March-Diaz R, Reyes JC (2008) The PHD domain of plant PIAS proteins mediates sumoylation of bromodomain GTE proteins. J Biol Chem 283:21469–21477. doi:10.1074/jbc.M708176200
Elrouby N, Coupland G (2010) Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA 107:17415–17420. doi:10.1073/pnas.1005452107
Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I (2007) NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19:1537–1548. doi:10.1105/tpc.106.049239
Matarasso N, Schuster S, Avni A (2005) A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant Cell 17:1205–1216. doi:10.1105/tpc.105.030775
Nigam N, Singh A, Sahi C, Chandramouli A, Grover A (2008) SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa L.) genes: genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol Genet Genomics 279:371–383. doi:10.1007/s00438-008-0318-5
Geisler-Lee J, O’Toole N, Ammar R, Provart NJ, Millar AH, Geisler M (2007) A predicted interactome for Arabidopsis. Plant Physiol 145:317–329. doi:10.1104/pp.107.103465
Supek F, Bosnjak M, Skunca N, Smuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. doi:10.1371/journal.pone.0021800
Richter K, Haslbeck M, Buchner J (2010) The heat shock response: life on the verge of death. Mol Cell 40:253–266. doi:10.1016/j.molcel.2010.10.006
Kotak S, Larkindale J, Lee U, von Koskull-Doring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10:310–316. doi:10.1016/j.pbi.2007.04.011
Mayer MP, Bukau B (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62:670–684. doi:10.1007/s00018-004-4464-6
Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143:251–262. doi:10.1104/pp.106.091322
Anckar J, Hietakangas V, Denessiouk K, Thiele DJ, Johnson MS, Sistonen L (2006) Inhibition of DNA binding by differential sumoylation of heat shock factors. Mol Cell Biol 26:955–964. doi:10.1128/MCB.26.3.955-964.2006
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Verslues PE, Juenger TE (2011) Drought, metabolites, and Arabidopsis natural variation: a promising combination for understanding adaptation to water-limited environments. Curr Opin Plant Biol 14:240–245. doi:10.1016/j.pbi.2011.04.006
Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20:1693–1707. doi:10.1105/tpc.107.057380
Chaikam V, Karlson DT (2010) Response and transcriptional regulation of rice SUMOylation system during development and stress conditions. BMB Rep 43:103–109
Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126:467–475. doi:10.1016/j.cell.2006.05.050
Lois LM, Lima CD, Chua NH (2003) Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15:1347–1359
Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21:3029–3038. doi:10.1093/emboj/cdf316
Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P (2005) A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 138:2061–2074. doi:10.1104/pp.105.060061
Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15:2122–2133. doi:10.1101/gad.204401
Nilsson L, Muller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30:1499–1512. doi:10.1111/j.1365-3040.2007.01734.x
Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39:792–796. doi:10.1038/ng2041
Wang X, Du G, Meng Y, Li Y, Wu P, Yi K (2010) The function of LPR1 is controlled by an element in the promoter and is independent of SUMO E3 Ligase SIZ1 in response to low Pi stress in Arabidopsis thaliana. Plant Cell Physiol 51:380–394. doi:10.1093/pcp/pcq004
Cuypers A, Smeets K, Ruytinx J, Opdenakker K, Keunen E, Remans T, Horemans N, Vanhoudt N, Van Sanden S, Van Belleghem F, Guisez Y, Colpaert J, Vangronsveld J (2011) The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. J Plant Physiol 168:309–316. doi:10.1016/j.jplph.2010.07.010
Curie C, Cassin G, Couch D, Divol F, Higuchi K, Le Jean M, Misson J, Schikora A, Czernic P, Mari S (2009) Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot 103:1–11. doi:10.1093/aob/mcn207
Hirayama T, Shinozaki K (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61:1041–1052. doi:10.1111/j.1365-313X.2010.04124.x
Qin F, Shinozaki K, Yamaguchi-Shinozaki K (2011) Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol 52:1569–1582. doi:10.1093/pcp/pcr106
van den Burg HA, Takken FL (2009) Does chromatin remodeling mark systemic acquired resistance? Trends Plant Sci 14:286–294. doi:10.1016/j.tplants.2009.02.003
van den Burg HA, Takken FL (2010) SUMO-, MAPK-, and resistance protein-signaling converge at transcription complexes that regulate plant innate immunity. Plant Signal Behav 5:1597–1601
Kim JM, To TK, Nishioka T, Seki M (2010) Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ 33:604–611. doi:10.1111/j.1365-3040.2009.02076.x
Meier I (2012) mRNA export and sumoylation-Lessons from plants. Biochim Biophys Acta 1819:531–537. doi:10.1016/j.bbagrm.2012.01.006
Muthuswamy S, Meier I (2011) Genetic and environmental changes in SUMO homeostasis lead to nuclear mRNA retention in plants. Planta 233:201–208. doi:10.1007/s00425-010-1278-7
Bossis G, Melchior F (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell 21:349–357. doi:10.1016/j.molcel.2005.12.019
Xu Z, Chan HY, Lam WL, Lam KH, Lam LS, Ng TB, Au SW (2009) SUMO proteases: redox regulation and biological consequences. Antioxid Redox Signal 11:1453–1484. doi:10.1089/ARS.2008.2182
Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F (2011) ROS signaling: the new wave? Trends Plant Sci 16:300–309. doi:10.1016/j.tplants.2011.03.007
Kim MG (2010) Alerted defense system attenuates hypersensitive response-associated cell death in arabidopsis siz1 mutant. J Plant Biol 53:70–78. doi:10.1007/s12374-009-9089-8
Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G (2010) Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J Exp Bot 61:4197–4220. doi:10.1093/jxb/erq282
Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R (2007) Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol 144:1777–1785. doi:10.1104/pp.107.101436
Tempe D, Piechaczyk M, Bossis G (2008) SUMO under stress. Biochem Soc Trans 36:874–878. doi:10.1042/BST0360874
Reeves PH, Murtas G, Dash S, Coupland G (2002) Early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129:5349–5361
Hermkes R, Fu YF, Nurrenberg K, Budhiraja R, Schmelzer E, Elrouby N, Dohmen RJ, Bachmair A, Coupland G (2011) Distinct roles for Arabidopsis SUMO protease ESD4 and its closest homolog ELS1. Planta 233:63–73. doi:10.1007/s00425-010-1281-z
Conti L, Kioumourtzoglou D, O’Donnell E, Dominy P, Sadanandom A (2009) OTS1 and OTS2 SUMO proteases link plant development and survival under salt stress. Plant Signal Behav 4:225–227
Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, Coruzzi GM, Gutierrez RA (2010) VirtualPlant: a software platform to support systems biology research. Plant Physiol 152:500–515. doi:10.1104/pp.109.147025
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Castro, P.H., Tavares, R.M., Bejarano, E.R. et al. SUMO, a heavyweight player in plant abiotic stress responses. Cell. Mol. Life Sci. 69, 3269–3283 (2012). https://doi.org/10.1007/s00018-012-1094-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-012-1094-2