Abstract
Periodontitis is a multifactorial infection characterized by inflammation and destruction of tooth supporting tissues, as a result of the response of a susceptible host to bacterial challenge. Studies have demonstrated that epigenetic events are able to influence the production of cytokines, contributing to the development of inflammatory diseases. Epigenetic events act through the remodeling of chromatin and can selectively activate or inactivate genes, determining their expression. The epigenetic process, by inducing a change in cytokine profile, may subsequently influence the pathogenesis and determine the outcome of many infectious diseases. These findings may have relevance for inflammatory diseases in which the expression of cytokines is unregulated. The purpose of this review is to show evidence that supports the hypothesis that epigenetic alterations, such as hyper and hypomethylation, of cytokine genes, could help to understand the mechanisms related to periodontal disease activity. Therefore, epigenetics may have future impact on diagnosis and/or therapeutics of periodontal disease.
Similar content being viewed by others
Introduction
Periodontitis is a multifactorial polymicrobial infection initiated by the presence of Gram-negative bacteria, which accumulates in the gingival crevice region [1]. The presence of periodontopathogenic bacteria leads to the establishment of an inflammatory process in periodontal tissues, which may lead to destruction of periodontal ligament and the adjacent supporting bone, causing tooth loss [2, 3].
Periodontal destruction is considered as a result of the response of a susceptible host to bacterial challenge [4]. The host’s response to infection depends on the nature and virulence of the pathogen, and is influenced by environmental and genetic factors [4, 5]. Periodontal disease is characterized by the intermittent occurrence of periods of activity, interrupted by periods of disease inactivity [6]. During disease activity, a burst of periodontal destruction mediated by cytokines is observed. Studies addressing periodontal disease activity and DNA methylation status of different cytokine genes will certainly add critical information concerning periodontal disease pathogenesis.
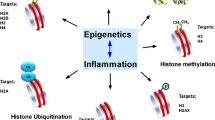
Epigenetics is described as changes in patterns of gene expression, which do not involve changes in the DNA sequence [7]. Epigenetic events act through chemical modifications of DNA and its associated proteins, thereby remodeling the chromatin and selectively activating or inactivating genes and determining their expression during development [7, 8].
DNA methylation and histone deacetylation are the two major mechanisms of epigenetic alterations observed in human cells [9]. DNA methylation is characterized by the addition of the methyl group onto cytosines within CpG regions. Histone deacetylation is a mechanism that involves the removal of the acetyl group leading to alteration of charge and packing of DNA around histones [10]. Both mechanisms block the binding of transcription factors. However, the histone modifications are considered more transient, whereas DNA methylation provides a stable form of gene regulation [11].
Recently, studies have demonstrated that epigenetic events are able to influence the production of cytokines, contributing to the development of diseases, such as airway inflammation and severe systemic inflammation [7, 12, 13]. However, the influence of epigenetics on cytokine production in periodontal disease has not yet been a matter of investigation. Therefore, the purpose of the present review is to show evidence that hyper and hypomethylation in cytokine genes could be involved in the development of periodontitis.
Epigenetic events and cytokines
DNA methylation is an active process catalyzed by DNA methyltransferases (DNMT) [14]. These enzymes have the ability to add methyl groups to the C5 position of DNA cytosines in CpG dinucleotides. CpG-rich areas of the DNA are known as CpG islands and are found mainly in the promoter region of genes [14]. Approximately, 50% of human genes contain CpG islands and most of these islands are unmethylated in normal tissue [14].
Unmethylated CpG islands are related to transcriptionally active structure, whereas methylated DNA recruits methyl-binding proteins, which promotes interaction with histone deacetylases (HDACs). HDAC remove acetyl groups of histones leading to chromatin compaction and preventing the binding of transcription factors [15–17]. Thus, gene products are not expressed and many cellular functions may be altered by DNA methylation, including DNA repair, cell proliferation regulation and inflammatory gene expression [7].
DNA methylation is maintained by DNMT1 and de novo methylation is mediated by DNMT3a and DNMT3b [14]. DNTM1 is associated with HDAC activity, indicating a cross-talk between these enzymes [18]. Both enzymes make up a transcriptional repression complex, being directly connected by methyl-binding protein. Based on this interaction, the use of DNMT1 and HDAC inhibitors have been suggested as therapeutic tools in chronic inflammation [11, 18].
In general, the expression of inflammatory pathways is influenced by epigenetic events since the recruitment of NF-kappaB requires the modification and remodeling of nucleosomes [19]. It has been suggested that cytokine genes are targets of multiple epigenetic events including demethylation-associated activation, monoallelic expression, cell cycle-associated regulation and methylation [13]. DNA methylation, for example, occurs in CD4+T lymphocytes when they differentiate into effectors Th1 and Th2 cells [10], which secrete predominantly IFN- γ or IL-4, respectively. During this process, the expression of one cytokine gene and the silencing of the other is regulated by epigenetic mechanisms [10] demonstrating that Th1/Th2 differentiation is dependent on epigenetic pathways of activation and silencing [19].
Epigenetic mechanisms have been evaluated in some cytokine genes as IL-2, IFN-γ, IL-10, IL-6, TNF-α and others [7, 13, 20–22]. Agents that inhibit DNA methylation induced an increase in the expression of the IL-2 gene [13]. It has been suggested that the expression of IFN-γ gene in humans is regulated by the status of methylation in its promoter region [20]. Recent evidence also indicates that changes related to methylation patterns can occur in the IL-10 gene during T cell differentiation into Th2 cells and in IL-10 producing regulatory T cells [7]. Other findings suggest that the hypermethylation of the IL-6 gene is, in part, produced by the repression of the gene [21]. It was demonstrated that epigenetic modifications of TNF-α gene actively regulate its expression and is present both constitutively and in response to acute stimulation of cells from the myeloid lineage [22]. These findings indicate that epigenetic changes occur in cytokine genes in human cells, affecting the ability of the cell to express cytokines.
Interestingly, it has been demonstrated that cytokines themselves might influence epigenetic changes in cells. IL-1 beta appears to provoke a marked repression of genes through activation of DNA methyltransferase [23]. Other data suggest that IL-6 might induce epigenetic events via regulation of the promoter region of DNA methyltransferase gene [24]. Wehbe et al. reported that IL-6 overexpression is related to altered expression and methylation of several genes, such as the epidermal growth factor receptor gene [25]. The overexpression of IL-6 may influence the expression and activity of DNMT, demethylases or histone expression, which participates in regulation of gene methylation [25].
The epigenetic process, by inducing a change in cytokine profile, may subsequently influence the pathogenesis and determine the outcome of many infectious diseases [10, 26]. These findings may have relevance for inflammatory diseases in which the expression of cytokines is unregulated.
Methylation of cytokine genes in human diseases
In human diseases, the majority of studies have evaluated the methylation pattern in cancer, in which epigenetic silencing has been associated to with tumor suppressor genes, DNA repair genes and other pathways critical to the neoplasic phenotype [9, 27]. However, epigenetic influence in other diseases is poorly known. With the purpose of introducing a new dimension to the biological significance of epigenetic events, studies of methylation pattern in cytokine genes in autoimmune and inflammatory diseases have been suggested [26].
Epigenetic investigations involving cytokine genes have been performed in some diseases, as in bronchial asthma [28] and in systemic lupus erythematosus [29]. In bronchial asthma, the degree of unmethylation of the IL-4 gene was higher in sensitized hosts and the extent of unmethylation correlated with IL-4 concentration [28]. In systemic lupus erythematosus, the hypomethylation of IL-4 and IL-6 promoters in activated T cells was observed, suggesting that T cells undergo demethylation during disease development [29].
Epigenetic control plays a major role in the inflammatory process, locally as well as systemically [12]. Evidence that epigenetic regulation is involved in rheumatoid arthritis [19, 30], airway inflammation [7] and during endotoxin tolerance [12] has also been suggested. In rheumatoid arthritis, the frequency of methylation in IL-10 gene was found to be higher when compared to healthy controls [30]. A recent study suggests that epigenetic investigation may raise the prospect of new treatment for inflammatory airway diseases where conventional therapy is not effective [7]. Also, epigenetic changes at the promoter region of proinflammatory genes mediate their repression during severe systemic inflammation [12].
Evidence is emerging to suggest that epigenetic events may be important in understanding the cause of interindividual variations in the inflammatory response [31]. Despite the small number of studies that evaluated epigenetic events in cytokine genes, the initial findings have offered insights into the pathogenesis of some diseases and therefore may contribute to the understanding of the development and severity of periodontal disease.
Epigenetics and periodontal disease
Studies evaluating the methylation pattern of cytokine genes may have relevance for inflammatory diseases in which the expression of some cytokines is altered. A complex network of pro- and anti-inflammatory cytokines act on the inflamed periodontal tissues, such as IL-1 beta, IL-1 alpha, TNF-alpha, IL-6, IL-10 and IL-4, and an overexpression of inflammatory cytokines has been demonstrated in individuals with periodontal disease [1, 2, 32].
Epigenetic events, such as hypomethylation and histone acetylation, are often associated with the innapropriate over-transcription of genes [29] and may be important in the pathogenesis of periodontal disease. Preliminary findings suggested that IL-6 gene is hypomethylated in tissues of individuals with periodontal disease compared to control samples, suggesting an overexpression of this cytokine in inflamed tissues [33].
Additionally, the overexpression of IL-6 might exert an epigenetic influence in cells by regulating the DNMT gene or by maintaining its methylation status, as previously shown in in vitro studies [24, 34, 35]. Stenvinkel et al. [34] speculated that a persistent inflammation may cause DNA methylation, which inactivates suppressors of cytokine signaling and contributes to exaggerated cytokine signaling. These findings are important since IL-6 is a key cytokine involved in bone resorption and has been detected in high levels in individuals with severe periodontitis [32].
Evidence also suggests that the presence of chronic inflammation and bacterial infection promotes DNA methylation [34, 36]. In this context, the potential for oral bacteria to alter DNA methylation patterns within the mucosa is currently under investigation [36]. However, it is not known if epigenetic mechanisms occur due to direct bacterial interaction with the tissue or as a consequence of a host inflammatory response [36].
Considering the findings above, epigenetic factors may interfere in the response of host to bacterial challenge and may contribute to the phenotype of periodontitis.
Factors that modify epigenetic mechanisms and periodontal disease
A study with monozygotic twins showed that patterns of DNA methylation were very similar in young twins, but in older twins the patterns were altered [37]. These data suggest that environmental factors present in the lifestyle of each individual induced the occurrence of epigenetic events that might influence the individual’s health [14].
Epigenome can be modified by several factors, such as toxic components of the environment, nutrition, tobacco smoke, alcohol and different infectious agents [14, 38]. Associations were observed between smoking and global DNA methylation and linked with a poor prognosis in lung cancer [39, 40]. As smoking is an important risk factor for periodontal disease, the evaluation of methylation in smokers with periodontitis may be of interest.
Additionally, age is a factor involved in an increase of methylation rate. In older individuals, the increase of gene silencing could contribute to the development of chronic diseases [19]. A previous study demonstrated that methylation of collagen-α1 gene in the periodontal ligament occurs during the aging process [41].
It is important to remember that DNA methylation regulates various biological functions in prokaryotes and eukaryotes. The DNA adenine methyltransferase (DAM) has been identified in oral bacteria and represents an important role in the control of the expression of virulent genes. Wu et al. [42] reported that DAM-mediated methylation may regulate genes required for the invasion process of A. actinomycetemcomitans.
As mentioned above, the maintenance of the methylation profile is influenced by several factors. However, the signaling pathways involved are not well known [43]. Although DNA methylation is recognized for its role in controlling many immune processes, the degree to which a gene undergoes transcriptional silencing will depend on both the level of cytosines that are methylated in the CpG islands and their positions within the sequence [14, 43].
Conclusion
Epigenetic mechanisms present in both oral pathogens or in the host could be important modulators of the immune response. Considering the complex and multifactorial nature of periodontal disease, it is possible to assume that epigenetic events could influence the establishment or progression of the disease. As epigenetic mechanisms have been demonstrated to impact on our understanding of inflammatory diseases, and given the scarce information of epigenetic events during periodontitis, it can be envisaged that this area of research will improve our knowledge of the pathogenesis of periodontitis.
The pathogenesis of periodontitis is complex. Genetic factors have been associated with an individual′s susceptibility to developing periodontitis [1]. Several genetic polymorphisms are associated with chronic periodontitis in different populations, mainly in cytokine genes [1, 5, 32]. However, the exposure of individuals to factors that influence the epigenome during their life may also alter the host response to bacterial presence. Genetic and epigenetic mechanisms are not mutually exclusive [14], but are likely to play coordinated roles in the regulating gene expression.
Studies evaluating the occurrence of epigenetic events in periodontal disease must be performed with caution. Periodontal disease has a multifactorial nature and several factors may influence both the epigenetic status and the periodontal conditions of individuals, such as smoking and age. The inclusion of a homogeneous population from the same geographical area, with similar socio-economic status and matched for confounding factors must be selected. A study with these characteristics should also consider the clinical parameters before associating it with epigenetic events. It can be envisaged that new diagnostic or therapeutic tools could be developed by the manipulation of the host and/or bacterial epigenome.
References
Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med. 2003;14(6):430–49.
Shapira L, Wilensky A, Kinane DF. Effect of genetic variability on the inflammatory response to periodontal infection. J Clin Periodontol. 2005;32:72–86.
Bodet C, Chandad F, Grenier D. Porphyromonas gingivalis-induced inflammatory mediator profile in an ex vivo human whole blood model. Clin Exp Immunol. 2006;143:50–7.
Borrell LN, Papapanou PN. Analytical epidemiology of periodontitis. J Clin Periodontol. 2005;32:132–58.
Takashiba S, Naruishi K. Gene polymorphisms in periodontal health and disease. Periodontol. 2006;2000(40):94–106.
American Academy of Periodontology. Epidemiology of periodontal disease. J Periodontol. 2005;76:1406–19.
Adcock IA, Tsaprouni L, Bhavsar P, Ito K. Epigenetic regulation of airway inflammation. Curr Opin Immunol. 2007;19:694–700.
Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–87.
Shaw R. The epigenetics of oral cancer. Int J Oral Maxillofac Surg. 2006;35:101–8.
Sanders VM. Epigenetic regulation of Th1 and Th2 cell development. Brain Behav Immun. 2006;20:317–24.
Bäckdahl L, Bushell A, Beck S. Inflammatory signalling as mediator of epigenetic modulation in tissue-specific chronic inflammation. Int J Biochem Cell Biol. 2009;41(1):176–84.
Gazzar ME, Yoza BK, Hu J, Cousart SL, McCall CE. Epigenetic silencing of tumor necrosis α during endotoxin tolerance. J Biol Chem. 2007;282:26857–64.
Fitzpatrick DR, Wilson CB. Methylation and demethylation in the regulation of genes, cells, and responses in the immune system. Clin Immunol. 2003;109:37–45.
Johnson IT, Belshaw NJ. Environment, diet and CpG island methylation: epigenetic signals in gastrointestinal neoplasia. Food Chem Toxicol. 2008;46:1346–59.
Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91.
Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9.
Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–63.
Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24(1):88–91.
Sánchez-Pernaute O, Ospelt C, Neidhart M, Gay S. Epigenetic clues to rheumatoid arthritis. J Autoimmun. 2008;30:12–20.
White GP, Watt PM, Holt BJ, Holt PG. Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO-T cells. J Immunol. 2002;168:2820–7.
Armenante F, Merola M, Furia A, Palmieri M. Repression of the IL-6 gene is associated with hypermethylation. Biochem Biophys Res Commun. 1999;258:644–7.
Sullivan KE, Reddy ABM, Dietzmann K, Suriano AR, Kocieda VP, Stewart M, et al. Epigenetic regulation of tumor necrosis factor alpha. Mol Cell Biol. 2007;27:5147–60.
Hmadcha A, Bedoya FJ, Sobrino F, Pintado E. Methylation-dependent gene silencing induced by interleukin 1b via nitric oxide production. J Exp Med. 1999;190:1595–603.
Hodge DR, Xiao W, Clausen PA, Heidecker G, Szyf M, Farrar WL. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erytroleukemia cells. J Biol Chem. 2001;276:39508–11.
Wehbe H, Henson R, Meng F, Mize-Berge J, Patel T. Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression. Cancer Res. 2006;66:10517–24.
Reiner SL. Epigenetic control in the immune response. Hum Mol Genet. 2005;14:R41–6.
Issa J-P. Age-related epigenetic changes and the immune system. Clin Immunol. 2003;109:103–8.
Kwon N-H, Kim J-S, Le J-Y, Oh M-J, Choi D-C. DNA methylation and the expression of IL-4 and IFN-g promoter genes in patients with bronchial asthma. J Clin Immunol. 2008;28:139–46.
Mi X, Zeng F. Hypomethylation of interleukin-4 and 6 promoters in T cells from systemic lupus erythematosus. Acta Pharmacol Sin. 2008;29:105–12.
Fu LH, Cong B, Zhen YF, Li SJ, Ma CL, Ni ZY, et al. Methylation status of the IL-10 gene promoter in the peripheral blood mononuclear cells of rheumatoid arthritis patients. Yi Chuan. 2007;29:1357–61. abstract.
Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol. 2008;79:1514–9.
Moreira PR, Lima PMA, Sathler KOB, Imanishi SA, Costa JE, Gomes RS, et al. Interleukin-6 expression and gene polymorphism are associated with severity of periodontal disease in a sample of Brazilian individuals. Clin Exp Immunol. 2007;148:119–26.
Offenbacher S, Barros SP, Beck JD. Rethinking periodontal inflammation. J Periodontol. 2008;79:1577–84.
Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, et al. Impact of inflammation on epigenetic DNA methylation: a novel risk factor for cardiovascular disease? J Intern Med. 2007;261:488–99.
Hodge DR, Peng B, Cherry JC, Hurt EM, Fox SD, Kelley JA, et al. Interleukin 6 supports the maintenance of p53 tumor suppressor gene promotor methylation. Cancer Res. 2005;65:4673–82.
Bobetsis YA, Barros SP, Lin DM, Weidman JR, Dolinoy DC, Jirtle RL, et al. Bacterial infection promotes DNA hypermethylation. J Dent Res. 2007;86:169–74.
Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–9.
Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–8.
Hillemacher T, Frieling H, Moskau S, Muschler MA, Semmler A, Kornhuber J, et al. Global DNA methylation is influenced by smoking behaviour. Eur Neuropsychopharmacol. 2008;18:295–8.
Kikuchi S, Yamada D, Fukami T, Maruyama T, Ito A, Asamura H, et al. Hypermethylation of the TSLC1/IGSF4 promoter is associated with tobacco smoking and a poor prognosis in primary non-small cell lung carcinoma. Cancer. 2006;106:1751–8.
Ohi T, Uehara Y, Takatsu M, Watanabe M, Ono T. Hypermethylation of CpG in the promoter of the COL1A1 gene in the aged periodontal ligament. J Dent Res. 2006;85:245–50.
Wu H, Lippmann JE, Oza JP, Zeng M, Fives-Taylor P, Reich NO. Inactivation of DNA adenine methyltransferase alters virulence factors in Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2006;21:238–44.
Bonilla-Henao V, Martinez R, Sobrino F, Pintado E. Different signaling pathways inhibit DNA methylation activity and up-regulate IFN-γ in human lymphocytes. J Leukoc Biol. 2005;78:1339–46.
Acknowledgments
RS Gomez, WO Dutra and PR Moreira are research fellows of Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: G. Wallace.
Rights and permissions
About this article
Cite this article
Gomez, R.S., Dutra, W.O. & Moreira, P.R. Epigenetics and periodontal disease: future perspectives. Inflamm. Res. 58, 625–629 (2009). https://doi.org/10.1007/s00011-009-0041-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-009-0041-7