Abstract
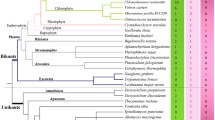
The MADS-box encodes a novel type of DNA-binding domain found so far in a diverse group of transcription factors from yeast, animals, and seed plants. Here, our first aim was to evaluate the primary structure of the MADS-box. Compilation of the 107 currently available MADS-domain sequences resulted in a signature which can strictly discriminate between genes possessing or lacking a MADS-domain and allowed a classification of MADS-domain proteins into several distinct subfamilies. A comprehensive phylogenetic analysis of known eukaryotic MADS-box genes, which is the first comprising animal as well as fungal and plant homologs, showed that the vast majority of subfamily members appear on distinct subtrees of phylogenetic trees, suggesting that subfamilies represent monophyletic gene clades and providing the proposed classification scheme with a sound evolutionary basis. A reconstruction of the history of the MADS-box gene subfamilies based on the taxonomic distribution of contemporary subfamily members revealed that each subfamily comprises highly conserved putative orthologs and recent paralogs. Some subfamilies must be very old (1,000 MY or more), while others are more recent. In general, subfamily members tend to share highly similar sequences, expression patterns, and related functions. The defined species distribution, specific function, and strong evolutionary conservation of the members of most subfamilies suggest that the establishment of different subfamilies was followed by rapid fixation and was thus highly advantageous during eukaryotic evolution. These gene subfamilies may have been essential prerequisites for the establishment of several complex eukaryotic body structures, such as muscles in animals and certain reproductive structures in higher plants, and of some signal transduction pathways. Phylogenetic trees indicate that after establishment of different subfamilies, additional gene duplications led to a further increase in the number of MADS-box genes. However, several molecular mechanisms of MADS-box gene diversification were used to a quite different extent during animal and plant evolution. Known plant MADS-domain sequences diverged much faster than those of animals, and gene duplication and sequence diversification were extensively used for the creation of new genes during plant evolution, resulting in a relatively large number of interacting genes. In contrast, the available data on animal genes suggest that increase in gene number was only moderate in the lineage leading to mammals, but in the case of MEF2-like gene products, heterodimerization between different splice variants may have increased the combinatorial possibilities of interactions considerably. These observations demonstrate that in metazoan and plant evolution, increased combinatorial possibilities of MADS-box gene product interactions correlated with the evolution of increasingly complex body plans.
Similar content being viewed by others
Abbreviations
- accession number:
-
(acc. no.)
- amino acids:
-
(aa)
- Genetics Computer Group:
-
(GCG)
- maximum parsimony method:
-
(MP)
- million years:
-
(MY)
- million years ago:
-
(MYA)
- neighbor-joining algorithm:
-
(NJ)
References
Affolter M, Montagne J, Walldorf U, Groppe J, Kloter U, LaRosa M, Gehring WJ (1994) TheDrosophila SRF homolog is expressed in a subset of tracheal cells and maps within a genomic region required for tracheal development. Development 120:743–753
Ainsworth C, Crossley S, Buchanan-Wollaston V, Thangavelu M, Parker J (1995) Male and female flowers of the dioecious plant sorrel show different patterns of MADS box gene expression. Plant Cell 7:1583–1598
Ammerer G (1990) Identification, purification, and cloning of a polypeptide (PRTF/GRM) that binds to mating-specific promoter elements in yeast. Genes Div 4:299–312
Angenent GC, Busscher M, Franken J, Mol JNM, van Tunen AJ (1992) Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell 4:983–993
Angenent GC, Franken J, Busscher M, Colombo L, van Tunen AJ (1993) Petal and stamen formation in petunia is regulated by the homeotic genefbp1. Plant J 4:101–112
Angenent GC, Franken J, Busscher M, Weiss D, van Tunen AJ (1994) Co-suppression of the petunia homeotic genefbp2 affects the identity of the generative meristem. Plant J 5:33–44
Angenent GC, Busscher M, Franken J, Dons HJM, van Tunen AJ (1995a) Functional interaction between the homeotic genesfbp1 andpMADS1 during petunia floral organogenesis. Plant Cell 7:507–516
Angenent GC, Franken J, Busscher M, van Dijken A, van Went JL, Dons HIM,van Tunen AJ (1995b) A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7: 1569–1582
Anthony RG, James PE, Jordan BR (1995) The cDNA sequence of a Cailiflowerapetala-1/squamosa homolog. Plant Physiol 108:441–442
Araki I, Satoh N (1995) As-MEF2 is predominantly expressed in mesenchyme and central nervous system in ascidian embryos. (unpublished)
Bairoch A (1991) PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res 19:2241–2245
Balavoine G, Telford MJ (1995) Identification of planarian homeobox sequences indicates the antiquity of most Hox/homeotic gene subclasses. Proc Natl Acad Sci USA 92:7227–7231
Baldauf SL, Palmer JD (1993) Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc Natl Acad Sci USA 90:11558–11562
Baudinette SC, Savin KW (1995) Carnation MADS-box genes. (unpublished)
Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development inArabidopsis thaliana byAPETALA1 and interacting genes. Development 119:721–743
Bradley D, Carpenter R, Sommer H, Hartley N, Coen E (1993) Complementary floral homeotic phenotypes result from opposite orientations of a transposon at theplena locus of Antirrhinum. Cell 72:85–95
Cacharrón J, Fischer A, Saedler H, Theißen G (1995) Expression patterns of MADS-box genes as studied byin situ hybridization. Maize Genet Coop Newslett 69:37–38
Carroll SB (1995) Homeotic genes and the evolution of arthropods and chordates. Nature 376:479–485
Chambers AE, Kotecha S, Towers N, Mohun TJ (1992) Muscle-specific expression of SRF-related genes in the early embryo ofXenopus leavis. EMBO J 11:4981–4991
Chung Y-Y, Kim S-R, Finkel D, Yanofsky MF, An G (1994) Early flowering and reduced apical dominance result from ectopic expression of a rice MADS box gene. Plant Mol Biol 26:657–665
Chung Y-Y, Kim S-R, Kang H-G, Noh Y-S, Park MC, Finkel D, An G (1995) Characterization of two rice MADS box genes homologous toGLOBOSA. Plant Sci 109:45–56
Colombo L, Franken J, Koetje E, van Went J, Dons HIM, Angenent GC, van Tunen AJ (1995) The petunia MADS box geneFBP11 determines ovule identity. Plant Cell 7:1859–1868
Davidson EH, Peterson KJ, Cameron RA (1995) Origin of bilaterian body plans: evolution of developmental regulatory mechanisms. Science 270:1319–1325
Davies B, Schwarz-Sommer Z (1994) Control of floral organ identity by homeotic MADS-box transcription factors. In: Nover L (ed) Results and problems in cell differentiation 20, Plant promoters and transcription factors. Springer, Berlin, pp 235–258
Dayhoff MO (1979) Atlas of protein sequences and structure, vol 5, supplement 3, 1978. National Biomedical Research Foundation, Washington, DC
Démolis N, Mallet L, Jacquet M (1994) A 12.5 kb fragment of the yeast chromosome II contains two adjacent genes encoding ribosomal proteins and six putative new genes, one of which encodes a putative transcriptional factor. Yeast 10:1511–1525
Dodou E, Sparrow DB, Mohun T, Treisman R (1995) MEF2 proteins, including MEF2A, are expressed in both muscle and non-muscle cells. Nucleic Acids Res 23:4267–4274
Doyle JJ (1994) Evolution of a plant homeotic multigene family: toward connecting molecular systematics and molecular developmental genetics. Syst Biol 43:307–328
Dubois E, Bercy J, Messenguy F (1987) Characterization of two genes,ARGRI andARGRIII, required for specific regulation of arginine metabolism in yeast. Mol Gen Genet 207:142–148
Edwards D, Feehan J (1980) Records ofCooksonia-type sporangia from late Wenlock strata in Ireland. Nature 287:41–42
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the boostrap. Evolution 39:783–791
Felsenstein J (1993) PHYLIP Phylogeny Inference Package 3.5. Department of Genetics, The University of Washington
Fischer A, Baum N, Saedler H, Theißen G (1995a) Chromosomal mapping of the MADS-box multigene family inZea mays reveals dispersed distribution of allelic genes as well as transposed copies. Nucleic Acids Res 23:1901–1911
Fischer A, Saedler H, Theissen G (1995b) Restriction fragment length polymorphism-coupled domain-directed differential display: a highly efficient technique for expression analysis of multigene families. Proc Natl Acad Sci USA 92:5331–5335
Fitch WW (1971) Toward defining the course of evolution: minimum change for a specified tree topology. Syst Zool 20:406–416
Flanagan CA, Ma H (1994) Spatially and temporally regulated expression of the MADS-box geneAGL2 in wild-type and mutantArabidopsis flowers. Plant Mol Biol 26:581–595
Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, Fritchman JL, Weidman JF, Small KV, Sandusky M, Fuhrmann J, Nguyen D, Utterback TR, Saudek DM, Phillips CA, Merrick JM, Tomb J-F, Dougherty BA, Bon KF, Hu P-C, Lucier TS, Peterson SN, Smith HO, Hutchison III CA, Venter JC (1995) The minimal gene complement ofMycoplasma geuitalium. Science 270:397–403
Garcia-Maroto F, Salamini F, Rohde W (1993) Molecular cloning and expression patterns of three alleles of theDeficiens-homologous geneSt-Deficiens fromSolanum tuberosum. Plant J 4:771–780
Gehring WJ (1992) The homeobox in perspective. Trends Biochem Sci 17:277–280
Gehring WJ, Affolter M, Biirglin T (1994) Homeodomain proteins. Annu Rev Biochem 63:487–526
Genetics Computer Group (1994) Program Manual for the Wisconsin Package, Version 8, September 1994, Genetics Computer Group, 575 Science Drive, Madison, Wisconsin, USA 53711
Gensel PG, Andrews HN (1984) Plant life in the Devonian. Praeger, New York
Gogarten JP (1994) Which is the most conserved group of proteins? Homology-orthology, paralogy, xenology, and the fusion of independent lineages. J Mol Evol 39:541–543
Gojobori T, Ikeo K (1994) Molecular evolution of serine protease and its inhibitor with special reference to domain evolution. Philos Trans R Soc Lond Biol 344:411–415
Goto K, Meyerowitz EM (1994) Function and regulation of theArabidopsis floral homeotic genePISTILLATA. Genes Dev 8:1548–1560
Hansen G, Estruch JJ, Sommer H, Spena A (1993) NTGLO: a tobacco homologue of theGLOBOSA floral homeotic gene ofAntirrhinum majus: cDNA sequence and expression pattern. Mol Gen Genet 239:310–312
Hardenack S, Ye D, Saedler H, Grant S (1994) Comparison of MADS box gene expression in developing male and female flowers of the dioecious plant white campion. Plant Cell 6:1775–1787
Hartland WB, Cox AV, Llewellyn PG, Pickton CAG, Smith AG, Walters R (1982) A geologic time scale. Cambridge University Press, Cambridge
Heard J, Dunn K (1995) Symbiotic induction of a MADS-box gene during development of alfalfa root nodules. Proc Natl Acad Sci USA 92:5273–5277
Heck GR, Perry SE, Nichols KW, Fernandez DE (1995) AGL15, a MADS domain protein expressed in developing embryos. Plant Cell 7:1271–1282
Hill CS, Wynne J, Treisman R (1995) The Rho family GTPases RhoA, Racl, and CDC42Hs regulate transcriptional activation by SRF. Cell 81:1159–1170
Hobson GM, Krahe R, Garcia E, Siciliano MJ, Funanage VL (1995) Regional chromosomal assignments for four members of the MADS domain transcription enhancer factor 2 (MEF2) gene family to human chromosomes 15826, 19p12, 5gl4, and lgl2-q23. Genomics 29:704–711
Holland PWH (1992) Homeobox genes in vertebrate evolution. Bioessays 14:267–273
Huang H, Mizukami Y, Hu Y, Ma H (1993) Isolation and characterization of the binding sequences for the product of theArabidopsis floral homeotic geneAGAMOUS. Nucleic Acids Res 21:4769–4776
Huang H, Tudor M, Weiss CA, Hu Y, Ma H (1995) TheArabidopsis MADS-box geneAGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol Biol 28:549–567
Huijser P, Klein J, Lönnig W-E, Meijer H. Saedler H, Sommer H (1992) Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box genesquamosa inAntirrhinum majus. EMBO J 11:1239–1249
Ingram GC, Goodrich J, Wilkinson MD, Simon R, Haughn OW, Coen ES (1995) Parallels betweenUNUSUAL FLORAL ORGANS andFIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell 7:1501–1510
Irish VF, Yamamoto YT (1995) Conservation of floral homeotic gene function between Arabidopsis and Antirrhinum. Plant Cell 7:1635–1644
Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic geneAPETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68:683–697
Jack T, Fox GL, Meyerowitz EM (1994) Arabidopsis homeotic geneAPETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76:703–716
Jarvis EE, Clark KL, Sprague GF (1989) The yeast transcription activator PRTF, a homolog of the mammalian serum response factor, is encoded by theMCM1 gene. Genes Dev 3:936–945
Jofuku KD, den Boer BGW, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic geneAPETALA2. Plant Cell 6:1211–1225
Johnson AD (1995) Molecular mechanisms of cell-type determination in budding yeast. Curr Opin Gen Dev 5:552–558
Kang S-G, Hannapel DJ (1995) Nucleotide sequences of novel potato (Solanum tuberosum L.)MADS-box cDNAs and their expression in vegetative organs. Gene 166:329–330
Kang H-G, Noh Y-S, Chung Y-Y, Costa MA, An K, An G (1995) Phenotypic alterations of petal and sepal by ectopic expression of a rice MADS box gene in tobacco. Plant Mol Biol 29:1–10
Kappen C, Ruddle FH (1993) Evolution of a regulatory gene family:HOM/HOX genes. Curr Opin Genet Dev 3:931–938
Kappen C, Schughart K, Ruddle FH (1989) Two steps in the evolution of Antennapedia-class vertebrate homeobox genes. Proc Natl Acad Sci USA 86:5459–5463
Kappen C, Schughart K, Ruddle FH (1993) Early evolutionary origin of major homeodomatn sequence classes. Genomics 18:54–70
Karunanandaa B, Kao T-H ( 1995) Characterization of a flower specific cDNA ofPetunia inflata encoding a putative homolog of Agamous protein. (unpublished)
Kaushal S, Schneider JW, Nadal-Ginard B. Mahdavi V (1994) Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science 266:1236–1240
Kempin SA, Mandel MA, Yanofsky MF (1993) Conversion of perianth into reproductive organs by ectopic expression of the tobacco floral homeotic geneNAG1. Plant Physiol 103:1041–1046
Kempin SA, Savidge B, Yanofsky MF (1995) Molecular basis of thecauliflower phenotype inArabidopsis. Science 267:522–525
Kim YS, Lee HS, Hoon LS, Yoo OJ, Jung WI, Liu JR (1995) The cDNA sequence of two MADS box genes inPanax ginseng (GenBank Z46612 for GAG2, Z46613 for GAGI) (PGR95-060). Plant Physiol 109:339
Knoll AH (1992) The early evolution of eukaryotes: a geological perspective. Science 256:622–627
Kobel HR, Du Pasquier L (1986) Genetics of polyploidXenopus. Trends Genet 2:310–315
Krause MW, Dichoso D, Park M, Fire A, Ahnn J (1995) Zygotic CeMEF-2 expression is not required for myogenesis in C. elegans. (unpublished)
Kush A, Brunelle A, Shevell D, Chua N-H (1993) The cDNA sequence of two MADS box proteins inPetunia. Plant Physiol 102:1051–1052
Laudet V, Stehelin D, Clevers H (1993) Ancestry and diversity of the HMG box superfamily. Nucleic Acids Res 21:2493–2501
Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM (1994) Isolation ofLuminidependens: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6:75–83
Leifer D, Krainc D, Yu Y-T, McDermott J, Breitbart RE, Heng J, Neve RL, Kosofsky B, Nadal-Ginard B, Lipton SA (1993) MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc Natl Acad Sci USA 90:1546–1550
Li W-H, Graur D (1991) Fundamentals of molecular evolution. Sinauer, Sunderland, MA
Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN (1994) D-MEF2; a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages duringDrosophila embryogenesis. Proc Natl Acad Sci USA 91:5662–5666
Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN (1995) Requirement of MADS domain transcription factor D-MEF2 for muscle formation inDrosophila. Science 267:688–693
Litman GW, Rast JP, Shamblott MJ, Haire RN, Hulst M, Roess W, Litman RT, Hinds-Frey KR, Zilch A, Amemiya CT (1993) Phylogenetic diversification of immunoglobulin genes and the antibody repertoire. Mol Biol Evol 10:60–72
Lönnig W-E, Saedler H (1994) The homeoticMacho mutant ofAntirrhinum majus reverts to wild-type or mutates to the homeoticplena phenotype. Mol Gen Genet 245:636–643
Lu Z-X, Wu M, Loh C-S, Yeong C-Y, Goh C-J (1993) Nucleotide sequence of a flower-specific MADS box cDNA clone from orchid. Plant Mol Biol 23:901–904
Ma H (1994) The unfolding drama of flower development: recent results from genetic and molecular analyses. Genes Dev 8:745–756
Ma H, Yanofsky MIT, Meyerowitz EM (1991)AGL1-AGL6, anArabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev 5:484–495
Mandel MA, Bowman JL, Kempin SA, Ma H, Meyerowitz EM, Yanofsky MF (1992a) Manipulation of flower structure in transgenic tobacco. Cell 71:133–143
Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992b) Molecular characterization of theArabidopsis floral homeotic geneAPETALA1. Nature 360:273–277
Mandel T, Lutziger I, Kuhlemeier C (1994) A ubiquitously expressed MADS-box gene fromNicotiana tabacum. Plant Mol Biol 25:319–321
Mandel MA, Yanofsky MF (1995) The ArabidopsisAGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated byAPETALA1. Plant Cell 7:1763–1771
Martin W, Lydiate D, Brinkmann H, Forkmann G, Saedler H, Cerff R (1993a) Molecular phylogenies in angiosperm evolution. Mol Biol Evol 10:140–162
Martin JF, Schwarz JJ, Olson EN (1993b) Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc Natl Acad Sci USA 90:5282–5286
Martin JF, Miano JM, Hustad CM, Copeland NG, Jenkins NA, Olson EN (1994) AMef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol Cell Biol 14:1647–1656
McDermott JC, Cardoso MC, Yu Y-T, Andres V, Leifer D, Krainc D, Lipton SA, Nadal-Ginard B (1993) hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol Cell Biol 13: 2564–2577
McGinnis W, Kuziora M (1994) The molecular architects of body design. Sci Am 270:36–42
Mena M, Mandel MA, Lerner DR, Yanofsky MF, Schmidt RJ (1995) A characterization of the MADS-box gene family in maize. Plant J 8:845–854
Menzel G, Apel K, Melzer S (1995) Isolation and analysis of SaMADS C, the APETALA 1 cDNA homolog from mustard. Plant Physiol 108:853–854
Meyerowitz EM (1994a) Flower development and evolution: new answers and new questions. Proc Natl Acad Sci USA 91:5735–5737
Meyerowitz EM (1994b) The genetics of flower development. Sci Am 271:40–47
Miller HG, Kocher TD, Loy B (1995) New MADS box domains inAsparagus officinalis L. Sex Plant Reprod 8:318–320
Mizukami Y, Ma H (1995) Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisenseAG RNA. Plant Mol Biol 28:767–784
Mohun TJ, Chambers AE, Towers N, Taylor MV (1991) Expression of genes encoding the transcription factor SRF during early development ofXenopus laevis: identification of a CArG box-binding activity as SRF. EMBO J 10:933–940
Montag K, Salamini F, Thompson RD (1995)ZEMa, a member of a novel group of MADS box genes, is alternatively spliced in maize endosperm. Nucleic Acids Res 23:2168–2177
Norman C, Runswick M, Pollock R, Treisman R (1988) Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55:989–1003
Nurrish SJ, Treisman R (1995) DNA binding specificity determinants in MADS-box transcription factors. Mol Cell Biol 15:4076–4085
Okada K, Shimura Y (1994) Genetic analyses of signalling in flower development usingArabidopsis. Plant Mol Biol 26:1357–1377
Olson EN, Perry M, Schulz RA (1995) Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol 172:2–14
Pabo CO, Sauer RT (1992) Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem 61:1053–1095
Passmore S, Maine GT, Elble R, Christ C, Tye B-K (1988)Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J Mol Biol 204:593–606
Pellegrini L, Tan S, Richmond TJ (1995) Structure of serum response factor core bound to DNA. Nature 376:490–498
Pendleton JW, Nagai BK, Murtha MT, Ruddle FH (1993) Expansion of theHox gene family and the evolution of chordates. Proc Natl Acad Sci USA 90:6300–6304
Philippe H, Chenuil A, Adoutte A (1994) Can the Cambrian explosion be inferred through molecular phylogeny? Dev Suppl: 15–25
Pnueli L, Abu-Abeid M, Zamir D, Nacken W, Schwarz-Sommer Z, Lifschitz E (1991) The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes fromAntirrhinum andArabidopsis. Plant J 1:255–266
Pnueli L, Hareven D, Broday L, Hurwitz C, Lifschitz E (1994a) TheTM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6:175–186
Pnueli L, Hareven D, Rounsley SD, Yanofsky MF, Lifschitz E (1994b) Isolation of the tomatoAGAMOUS geneTAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6:163–173
Pollock R, Treisman R (1991) Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev 5: 2327–2341
Pu WT, Struhl K (1991) Highly conserved residues in the bZIP domain of yeast GCN4 are not essential for DNA binding. Mol Cell Biol 11:4918–4926
Purugganan MD, Wessler SR (1994) Molecular evolution of the plantR regulatory gene family. Genetics 138:849–854
Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF (1995) Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics 140:345–356
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) TheCONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857
Reiser L, Modrusan Z, Margossian L, Samach A, Ohad N, Haughn GW, Fischer RL (1995) TheBELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83:735–742
Riley DE, Krieger JN (1995) Molecular and phylogenetic analysis of PCR-amplified cyclin-dependent kinase (CDK) family sequences from representatives of the earliest available lineages of eukaryotes. J Mol Evol 41:407–413
Rounsley SD, Ditta GS, Yanofsky MF (1995) Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7:1259–1269
Saedler H, Huijser P (1993) Molecular biology of flower development inAntirrhinum majus (snapdragon). Gene 135:239–243
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sakai H, Medrano LJ, Meyerowitz EM (1995) Role of SUPERMAN in maintainingArabidopsis floral whorl boundaries. Nature 378:199–203
Savard L, Li P, Strauss SH, Chase MW, Michaud M], Bousquet J (1994) Chloroplast and nuclear gene sequences indicate Late Pennsylvania time for the last common ancestor of extant seed plants. Proc Natl Acad Sci 91:5163–5167
Savidge B, Rounsley SD, Yanofsky MF (1995) Temporal relationship between transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell 7:721–733
Schena M, Davis RW (1994) Structure of homeobox-leucine zipper genes suggests a model for the evolution of gene families. Proc Natl Acad Sci USA 91:8393–8397
Schmidt RJ, Veit B, Mandel MA, Mena M, Hake S, Yanofsky MF (1993) Identification and molecular characterization ofZAG1, the maize homolog of the Arabidopsis floral homeotic geneAGAMOUS. Plant Cell 5:729–737
Schubert FR, Nieselt-Struwe K, Gross P (1993) The Antennapedia-type homeobox genes have evolved from three precursors separated early in metazoan evolution. Proc Natl Acad Sci USA 90:143–147
Schughart K, Kappen C, Ruddle FH (1989) Duplication of large genomic regions during the evolution of vertebrate homeobox genes. Proc Natl Acad Sci USA 86:7067–7071
Schwarz-Sommer Z, Huijser P. Nacken W, Saedler H, Sommer H (1990) Genetic control of flower development by homeotic genes inAntirrhinum majus. Science 250:931–936
Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönnig W-E, Saedler H, Sommer H (1992) Characterization of theAntirrhinum floral homeotic MADS-box genedeficiens; evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J 11:251–263
Scott MP (1993) A rational nomenclature for vertebrate homeobox (HOX) genes. Nucleic Acids Res 21:1687–1688
Shiraishi H, Okada K, Shimura Y (1993) Nucleotide sequences recognized by the AGAMOUS MADS domain ofArabidopsis thaliana in vitro. Plant J 4:385–398
Shore P, Sharrocks AD (1995) The MADS-box family of transcription factors. Eur J Biochem 229:1–13
Sommer H, Beltrán J-P, Huijser P, Pape H, Lönnig W-E, Saedler H, Schwarz-Sommer Z (1990)Deficiens, a homeotic gene involved in the control of flower morphogenesis inAntirrhinum majus: the protein shows homology to transcription factors. EMBO J 9:605–613
Strauss SH, Rottmann WH, Brunner AM, Sheppard LA (1995) Genetic engineering of reproductive sterility in forest trees. Mol Breeding 1:5–26
Tandre K, Albert VA, Sundas A, Engström P (1995) Conifer homologues to genes that control floral development in angiosperms. Plant Mol Biol 27:69–78
Theißen G, Saedler H (1995) MADS-box genes in plant ontogeny and phylogeny: Haeckel's ‘biogenefc law’ revisited. Curr Opin Genet Dev 5:628–639
Theißen G, Strater T, Fischer A, Saedler H (1995) Structural characterization, chromosomal localization and phylogenetic evaluation of two pairs ofAGAMOUS-likeMADS-box genes from maize. Gene 156:155–166
Treisman R (1990) The SRE: a growth factor responsive transcriptional regulator. Semin Cancer Biol 1:47–58
Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lönnig W-E, Saedler H, Sommer H, Schwarz-Sommer Z (1992)GLOBOSA: a homeotic gene which interacts withDEFICIENS in the control ofAntirrhinum floral organogenesis. EMBO J 11:4693–4704
Tsuchimoto S, van der Krol AR, Chua N-H (1993) Ectopic expression ofpMADS3 in transgenic petunia phenocopies the petuniablind mutant. Plant Cell 5:843–853
van der Krol AR, Chua N-H (1993) Flower development in petunia. Plant Cell 5:1195–1203
van der Krol AR, Brunelle A, Tsuchimoto S, Chua N-H (1993) Functional analysis of petunia floral homeotic MADS box genepMADS1. Genes Dev 7:1214–1228
Watanabe Y, Irie K, Matsumoto K (1995) YeastRLM1 encodes a serum response factor-like protein that may function downstream of the Mpkl (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol 15:5740–5749
Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78:203–209
Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992)LEAFY controls floral meristem identity in Arabidopsis. Cell 69:843–859
Wolfe KH, Gouy M, Yang Y-W, Sharp PM, Li W-H (1989) Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA 86:6201–6205
Wolffe AP (1994) Structural and functional properties of the evolutionary ancient Y-box family of nucleic acid binding proteins. Bioessays 16:245–251
Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by theArabidopsis homeotic geneagamous resembles transcription factors. Nature 346:35–39
Yu Y-T, Breitbart RE, Smoot LB, Youngsook L, Mahdavi V, Nadal-Ginard B (1992) Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev 6:1783–1798
Author information
Authors and Affiliations
Additional information
Correspondence to: G. Theißen
Rights and permissions
About this article
Cite this article
Theißen, G., Kim, J.T. & Saedler, H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol 43, 484–516 (1996). https://doi.org/10.1007/BF02337521
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02337521