Abstract
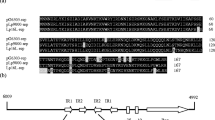
We have determined the nucleotide sequences of two structural genes of the Escherichia coli gab cluster, which encodes the enzymes of the 4-aminobutyrate degradation pathway: gabD, coding for succinic semialdehyde dehydrogenase (SSDH, EC 1.2.1.16) and gabP, coding for the 4-aminobutyrate (GABA) transport carrier (GABA permease). We have previously reported the nucleotide sequence of the third structural gene of the cluster, gabT, coding for glutamate: succinic semialdehyde transaminase (EC 2.6.1.19). All three gab genes are transribed unidirectionally and their orientation within the cluster is 5′-gabD-gabT-gabP-3′. gabT and gabP are separated by an intergenic region of 234-bp, which contains three repetetive extragenic palindromic (REP) sequences. The gabD gene consists of 1,449 nucleotides specifying a protein of 482 amino acids with a molecular mass of 51.7 kDa. The protein shows significant homologies to the NAD+-dependent aldehyde dehydrogenase (EC 1.2.1.3) from Aspergillus nidulans and several mammals, and to the tumor associated NADP+-dependent aldehyde dehydrogenase (EC 1.2.1.4) from rat. The permease gene gabP comprises 1,401 nucleotides coding a highly hydrophobic protein of 466 amino acids with a molecular mass of 51.1 kDa. The GABA permease shows features typical for an integral membrane protein and is highly homologous to the aromatic acid carrier from E. coli, the proline, arginine and histidine permeases from Saccharomyces cerevisiae and the proline transport protein from A. nidulans. Uptake of GABA was increased ca. 5-fold in transformants of E. coli containing gabP plasmids. Strong overexpression of the gabP gene under control of the isopropyl-2-d-thiogalactoside (IPTG) inducible tac promoter, however, resulted in a severe growth inhibition of the transformed strains. The GABA carrier was characterized using moderately overexpressing transformants. The K m of GABA uptake was found to be 11.8 μM and the Vmax 0.33 nmol/min · mg cells. Uptake of GABA was stimulated by ammonium sulfate and abolished by 2,4-dinitrophenol. Aspartate competed with GABA for uptake.
Similar content being viewed by others
References
Bartsch K, Dichmann R, Schmitt P, Uhlmann E, Schulz A (1990a) Stereospecific production of the herbicide phosphinothricin (glufosinate) by transamination: cloning, characterization, and overexpression of the gene encoding a phosphinothricin-specific transaminase from Escherichia coli. Appl Environ Microbiol 56: 7–12.
Bartsch K, Johnn-Marteville Avon, Schulz A (1990b) Molecular analysis of two genes of the Escherichia coli gab cluster: Nucleotide sequence of the glutamate: succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD). J Bacteriol 172: 7035–7042.
Bennetzen JL, Hall BD (1982) Codon selection in yeast J Biol Chem 257: 3026–3031.
Buchel DE, Gronenborn B, Müller-Hill B (1980) Sequence of the lactose permease gene. Nature 283: 541–545.
Chen EY, Seeburg PH (1985) Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA 4: 165–170.
Dover S, Halpern YS (1972a) Utilization of γ-aminobutyric acid as the sole carbon and nitrogen source by Escherichia coli K-12 mutants. J Bacteriol 109: 835–843.
Dover S, Halpern YS (1972b) Control of the pathway of γ-aminobutyrate breakdown in Escherichia coli K-12. J Bacteriol 110: 165–170.
Dover S, Halpern YS (1974) Genetic analysis of the γ-aminobutyrate utilization pathway in Escherichia coli K-12. J Bacteriol 117: 494–501.
Eya S, Maeda M, Tomochika KI, Kanemasa Y, Futai M (1989) Overproduction of truncated subunit a of H+-ATPase causes growth inhibition of Escherichia coli. J Bacteriol 171: 6853–6858.
Farres J, Guan KL, Weiner H (1989) Primary structures of rat and bovine liver mitochondrial aldehyde dehydrogenases deduced from cDNA sequences. Eur J Biochem 180: 67–74.
Fürste JP, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E (1986) Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48: 119–131.
Gilson E, Clément JM, Brutlag D, Hofnung M (1984) A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J 3: 1417–1421.
Henikoff S (1984) Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28: 351–359.
Hoffmann W (1985) Molecular characterization of the CAN1 locus in Saccharomyces cerevisiae. J Biol Chem 260: 11831–11837.
Honoré N, Cole ST (1990) Nucleotide sequence of the aroP gene encoding the general aromatic amino acid transport protein of Escherichia coli K-12: homology with yeast transport proteins. Nucleic Acids Res 18: 653.
Hsu LC, Bendel RE, Yoshida A (1988) Genomic structure of the human mitochondrial aldehyde dehydrogenase gene. Genomics 2: 57–65.
JonesJr DE, Brennan MD, Hempel J, Lindahl R (1988) Cloning and complete nucleotide sequence of a full-length cDNA encoding a catalytically functional tumor-associated aldehyde dehydrogenase. Proc Natl Acad Sci USA 85: 1782–1786.
Kahane S, Levitz R, Halpern YS (1978) Specificity and regulation of γ-aminobutyrate transport in Escherichia coli. J Bacteriol 135: 295–299.
Kalman M, Gentry DR, Cashel M (1991) Characterization of the Escherichia coli K-12 gltS glutamate permease gene. Mol Gen Genet 225: 379–386.
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132.
Levinson A, Silver D, Seed B (1984) Minimal size plasmids containing an M13 origin for production of single-strand transducing particles. J Mol Appl Genet 2: 507–517.
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Messing J, Crea R, Seeburg PH (1981) A system for shotgun DNA sequencing. Nucleic Acids Res 9: 309–313.
Metzer E, Halpern Y (1990) In vivo cloning and characterization of the gabCTDP gene cluster of Escherichia coli K-12. J Bacteriol 172: 3250–3256.
Moryama Y, Maeda M, Futai M (1990) Energy coupling of l-glutamate transport and vacuolar H+-ATPase in brain synaptic vesicles. J Biochem 108: 689–693.
Pickett M, Gwynne DI, Buxton FP, Elliott R, Davies RW. Lockington RA, Scazzocchio C, Sealy-Lewis HM (1987) Cloning and characterization of the aldA gene of Aspergillus nidulans. Gene 51: 217–226.
Saier MH, Werner PK, Müller M (1989) Insertion of proteins into bacterial membranes: mechanism, characteristics, and comparisons with the eucaryotic process. Microbiol Rev 53: 333–366.
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5468.
Schulz A, Taggeselle P, Tripier D, Bartsch K (1990) Stereospecific production of the herbicide phosphinothricin (glufosinate) by transamination: isolation and characterization of a phosphinothricin-specific transaminase from Escherichia coli. Appl Environ Microbiol 56: 1–6.
Sophianopoulou V, Scazzocchio C (1989) The proline transport protein of Aspergillus nidulans is very similar to amino acid transporters of Saccharomyces cerevisiae. Mol Microbiol 3: 705–714.
Stern MJ, Ferro-Luzzi Ames G, Smith NH, Robinson EC, Higgins CF (1984) Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell 37: 1015–1026.
Tanaka J, Fink GR (1985) The histidine permease gene (HIP1) of Saccharomyces cerevisiae. Gene 38: 205–214.
Urdea MS, Merryweather JP, Mullenbach GT, Coid D, Heberlein U, Valenzuela P, Barr J (1983) Chemical synthesis of a gene for human epidermal growth factor urogastrone and its expression in yeast. Proc Natl Acad Sci USA 80: 7461–7465.
Vandenbol M, Jauniaux JC, Grenson M (1989) Nucleotide sequence of the Saccharomyces cerevisiae PUT4 proline-permease-encoding gene: similarities between CAN1, HIP1 and PUT4 permeases. Gene 83: 153–159.
Wallace B, Yang YJ, Hong J, Lum D (1990) Cloning and sequencing of a gene encoding a glutamate and aspartate carrier of Escherichia coli K-12. J Bacteriol 172: 3214–3220.
Zaboura M, Halpern YS (1978) Regulation of γ-aminobutyric acid degradation in Escherichia coli by nitrogen metabolism enzymes. J Bacteriol 133: 447–451.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Niegemann, E., Schulz, A. & Bartsch, K. Molecular organization of the Escherichia coli gab cluster: nucleotide sequence of the structural genes gabD and gabP and expression of the GABA permease gene. Arch. Microbiol. 160, 454–460 (1993). https://doi.org/10.1007/BF00245306
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00245306