Abstract
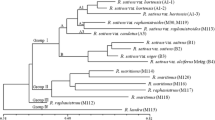
Restriction fragment length polymorphism (RFLP) and random amplified polymorphic DNA (RAPD) markers are being used widely for evaluating genetic relationships of crop germplasm. Differences in the properties of these two markers could result in different estimates of genetic relationships among some accessions. Nuclear RFLP markers detected by genomic DNA and cDNA clones and RAPD markers were compared for evaluating genetic relationships among 18 accessions from six cultivated Brassica species and one accession from Raphanus sativus. Based on comparisons of genetic-similarity matrices and cophenetic values, RAPD markers were very similar to RFLP markers for estimating intraspecific genetic relationships; however, the two marker types gave different results for interspecific genetic relationships. The presence of amplified mitochondrial and chloroplast DNA fragments in the RAPD data set did not appear to account for differences in RAPD- and RFLP-based dendrograms. However, hybridization tests of RAPD fragments with similar molecular weights demonstrated that some fragments, scored as identical, were not homologous. In all these cases, the differences occurred at the interspecific level. Our results suggest that RAPD data may be less reliable than RFLP data when estimating genetic relationships of accessions from more than one species.
Similar content being viewed by others
References
Demeke T, Adams RP, Chibbar R (1992) Potential taxonomic use of random amplified polymorphic DNA (RAPD): a case study in Brassica. Theor Appl Genet 84:990–994
Diers BW, Osborn TC (1994) Genetic diversity of oilseed Brassica napus germplasm based on restriction fragment length polymorphisms. Theor Appl Genet (in press)
Efron B, Tibishirani R (1986) Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat Sci 1:54–77
Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132:6–13
Gower JC (1985) Measures of similarity, dissimilarity, and distance. In: Kotz S, Johnson NL (eds) Encyclopedia of statistical sciences, vol 5. Wiley, New York, pp 397–405
Hadrys H, Balick M, Schierwater B (1992) Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol Ecol 1:55–63
Hu J, Quiros CF (1991) Identification of broccoli and cauliflower cultivars with RAPD markers. Plant Cell Rep 10:505–511
Kidwell KK, Woodfield DR, Bingham ET, Osborn TC (1994) Molecular marker diversity and yield of isogenic 2x and 4x single crosses of alfalfa. Crop Sci 34:784–788
Klein-Lankhorst RM, Vermunt A, Weide R, Liharska T, Zabel P (1991) Isolation of molecular markers for tomato (L. esculentum) using random amplified polymorphic DNA (RAPD). Theor Appl Genet 83:83–108
Lee M, Godshalk EB, Lamkey KR, Woodman WW (1989) Association of restriction fragment length polymorphisms among maize inbreds with agronomic performance of their crosses. Crop Sci 29:1067–1071
Maniatis M, Hardison RC, Lacy E, Lauer J, O'Connell C, Quon D, Sim DK, Efstradiatis A (1978) The isolation of structural genes from libraries of eukaryotic DNA. Cell 15:687–701
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Melchinger AE, Lee M, Lamkey KR, Hallauer AR, Woodmann WL (1990) Genetic diversity for restriction fragment length polymorphisms and heterosis for two diallel sets of maize inbreds. Theor Appl Genet 80:488–496
Miller JC, Tanksley SD (1990) RFLP analyses of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor Appl Genet 80:437–448
Murray M, Thompsom WF (1980) Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res 8:4321–4325
Neuhausen SL (1992) Evaluation of restriction fragment length polymorphism in Cucumis melo. Theor Appl Genet 83:379–384
Osborn TC, Alexander DC, Fobes JF (1987) Identification of restriction fragment length polymorphism linked to genes controlling soluble-solids content in tomato fruit. Theor Appl Genet 73:350–356
Perbal B (1984) A practical guide to molecular cloning. John Wiley and sons, New York
Reid RA, John MC, Amasino RM (1988) Deoxyribonuclease I sensitivity of the T-DNA ipt gene is associated with gene expression. Biochemistry 27:5748–5754
Rohlf FJ (1992) NTSYS-pc Numerical taxonomy and multivariate analysis system version 1.7. Owner Manual
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Smith JSC, Smith OS (1991) Restriction fragment length polymorphism can differentiate among U.S. maize hybrids. Crop Sci 31:893–899
Smith OS, Smith JSC, Bowen SL, Tenborg RA, Wall SJ (1990) Similarities among a group of elite maize inbreds as measured by pedigree, F1 grain yield, heterosis and RFLPs. Theor Appl Genet 80:833–840
Sneath PHA, Sokal RR (1973) Numerical taxonomy. W.H. Freeman, San Francisco
Song KM, Osborn TC, Williams PH (1988) Brassica taxonomy based on restriction fragment length polymorphisms (RFLPs). 1. Genome evolution of diploid and amphidiploid species. Theor Appl Genet 75:784–794
Song KM, Osborn TC, Williams PH (1990) Brassica taxonomy based on restriction fragment length polymorphisms (RFLPs). 3. Genome relationship in Brassica and related genera and the origin of B. oleracea and B. rapa (syn. campestris). Theor Appl Genet 79:497–506
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Tivang JG (1992) Sampling variance of molecular marker data using the bootstrap procedure. MS thesis, University of Wisconsin-Madison
Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18:7213–7218
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:7213–7218
Author information
Authors and Affiliations
Additional information
Communicated by A. L. Kahler
Rights and permissions
About this article
Cite this article
Thormann, C.E., Ferreira, M.E., Camargo, L.E.A. et al. Comparison of RFLP and RAPD markers to estimating genetic relationships within and among cruciferous species. Theoret. Appl. Genetics 88, 973–980 (1994). https://doi.org/10.1007/BF00220804
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00220804