Abstract
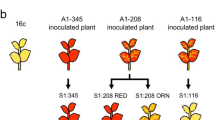
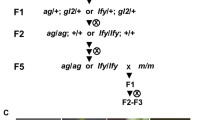
In several plant systems expression of structurally intact genes may be silenced epigenetically when a transgenic construct increases the copy number of DNA sequences. Here we report epigenetic silencing inArabidopsis lines containing transgenic inserts of defined genetic structure, all at the same genomic locus. These comprise an allelic series that includes a single copy of the primary insert, which carries repeated drug resistance transgenes, and a set of its derivatives, which as a result of recombination within the insert carry different numbers and alleles of resistance genes. Although the drug resistance genes remained intact, both the primary and some recombinant lines nevertheless segregated many progeny that were partly or fully drug-sensitive because of silencing. As in other systems silencing was reversible, and correlated with decreased steady-state mRNA and increased DNA methylation. Each different number and combination of genes, on the same or different (i.e., homologous) chromosomes, conditioned its own idiosyncratic segregation pattern. Strikingly, lines with a single gene segregated only a few slightly drug-sensitive progeny whereas multi-gene lines segregated many highly sensitive progeny, indicating dependence of silencing at this locus on repeated sequences. This argues strongly against explanations based on antisense RNA, but is consistent with explanations based on ectopic DNA pairing. One possibility is that silencing reflects the interaction of paired homologous DNA with flanking heterologous DNA, which induces condensation of chromatin into a non-transcribable state.
Similar content being viewed by others
References
Antequera F, Bird AP: Unmethylated CpG islands associated with genes in higher plant DNA. EMBO J 7: 2295–2299 (1988).
Aparicio OM, Billington BL, Gottschling DE: Modifiers of position effect are shared between telomeric and silent mating-type loci inS. cerevisiae. Cell 66: 1279–1287 (1991).
Assaad FF: Duplicated transgenes inArabidopsis: recombination and gene silencing. Ph.D. Thesis, Massachusetts Institute of Technology (1992).
Assaad FF, Signer ER: Cauliflower mosaic virus P35S promoter activity inEscherichia coli. Mol Gen Genet 223: 517–520 (1990).
Assaad FF, Signer ER: Somatic and germinal recombination of a direct repeat in Arabidopsis. Genetics 132: 553–566 (1992).
Banks JA, Masson P, Federoff N: Molecular mechanisms in the developmental regulation of the maizeSuppressor-mutator transposable element. Genes Devel 2: 1364–1380 (1988).
Brink RA: Paramutation and chromosome organization. Q Rev Biol 35: 120–137 (1960).
Brink RA: Paramutation. Annu Rev Genet 7: 129–152 (1973).
Cameron FH, Jennings PA: Inhibition of gene expression by a short sense fragment. Nucl Acids Res 19: 469–475 (1990).
Cedar H: DNA methylation and gene activity. Cell 53: 3–4 (1988).
Church G, Gilbert W: Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 (1984).
de Carvalho F, Gheysen G, Kushnir S Van Montagu M, Inze D, Castresana S: Suppression of β-1,3-glucanase transgene expression in homozygous plants. EMBO J 11: 2595–2602 (1992).
Dooner HK, Robbins TP, Jorgensen RA: Genetic and developmental control of anthocyanin biosynthesis. Annu Rev Genet 25: 173–199 (1991).
Elkind Y, Edwards R, Mevandad M, Hedrick SA, Ribak O, Dixon RA, Lamb CJ: Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc Natl Acad Sci USA 87: 9057–9061 (1990).
Faugeron G, Rhounim L, Rossignol JL: How does the cell count the number of ectopic copies of a gene in the premeitotic inactivation process acting inAscobolus immersus? Genetics 128: 585–591 (1990).
Feinberg A, Vogelstein B: A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 (1983).
Felsenfeld G: Chromatin as an essential part of the transcription mechanism. Nature 355: 219–224 (1992).
Goring DR, Thomson L, Rothstein S: Transformation of a partial nopaline synthase gene into tobacco suppresses the expression of a resident wild-type gene. Proc Natl Acad Sci USA 88: 1770–1774 (1991).
Grierson D, Fray RG, Hamilton AJ, Smith CJS, Watson CF: Does co-suppression of sense genes in transgenic plants involve antisense RNA? Trends Biotechnol 9: 122–123 (1991).
Henikoff S, Dreesen TD: Trans-inactivation of theDrosophila brown gene: evidence for transcriptional repression and somatic pairing dependence. Proc Natl Acad Sci USA 86: 6704–6708 (1989).
Heslop-Harrison JS, Bennett MD: Nuclear architecture in plants. Trends Genet 6: 401–405 (1991).
Hobbs SLA, Kpodar P, DeLong CMO: The effect of T-DNA copy number, position and methylation on reporter gene expression in tobacco transformants. Plant Mol Biol 15: 851–864 (1990).
Holliday R: The inheritance of epigenetic defects. Science 238: 163–170 (1987).
Howard-Flanders P, West SC, Stasiak A: Role of RecA protein spiral filaments in genetic recombination. Nature 309: 215–220 (1984).
Jones JDG, Gilbert DE, Grady KL, Jorgensen RA: T-DNA structure and gene expression in petunia plants transformed byAgrobacterium tumefaciens C58 derivatives. Mol Gen Genet 204: 478–485 (1987).
Jorgensen R: Altered gene expression in plants due totrans interactions between homologous genes. Trends Biotechnol 8: 340–344 (1990).
Jorgensen R: Beyond antisense — How do transgenes interact with homologous plant genes? Trends Biotechnol 9: 266–267 (1991).
Jorgensen R: Silencing of plant genes by homologous transgenes. AgBiotech News Inform 4: 265N-273N (1991).
Kricker MC, Drake JW, Radman M: Duplication-targeted DNA methylation and mutagenesis in evolution of eukaryotic chromosomes. Proc Natl Acad Sci USA 89: 1075–1079 (1992).
Linn F, Heidmann I, Saedler H, Meyer P: Epigenetic changes in the expression of the maize A1 gene inPetunia hybrida: role of numbers of integrated gene copies and state of methylation. Mol Gen Genet 222: 329–336 (1990).
Marks M, West J, Weeks D: The relatively large betatubulin family ofArabidopsis contains a member with an unusual transcribed 5′ noncoding sequence. Plant Mol Biol 10: 91–104 (1987).
Matzke MA, Matzke AJM: Gene interactions and epigenetic variation in transgenic plants. Devel Genet 11: 214–223 (1990).
Matzke MA, Primig M, Trnovsky J, Matzke AJM: Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J 8: 643–649 (1989).
Mittelsten Scheid O, Paszkowski J, Potrykus I: Reversible inactivation of a transgene inArabidopsis thaliana. Mol Gen Genet 228: 104–112 (1991).
Mol JNM, van Blokland R, Kooter J: More about cosuppression. Trends Biotechnol 9: 182–183 (1991).
Napoli C, Lemieux C, Jorgensen R: Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes inrans. Plant Cell 2: 279–289 (1990).
Pandit NN, Russo VEA: Reversible inactivation of a foreign gene,hph, during the asexual cycle inNeurospora crassa transformants. Mol Gen Genet 234: 412–422 (1992).
Pirrotta V: Transvection and long-distance gene regulation. BioEssays 12: 409–414 (1990).
Rand D: RIPping and RAPping at Berkeley. Genetics 132: 1123–1224 (1992).
Rhounim L, Rossignol JL, Faugeron G: Epimutation of repeated genes inAscobolus immersus. EMBO J 11: 4451–4457 (1992).
Robbins TP, Walker EL, Kermicle JL, Alleman M, Dellaporta SL: Meiotic instabiity of theR-r complex arising from displaced intragenic exchange and intrachromosomal rearrangement. Genetics 129: 271–283 (1991).
Rogers SO, Bendich AJ: Extraction of DNA from plant tissues. In: Dure L, Schilperoort RA (eds) Plant Molecular Biology Manual, pp. A6: 1–10. Kluwer Academic Publishers, Dordrecht (1988).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Selker EU: Premeiotic instability of repeated sequences inNeurospora crassa. Annu Rev Genet 24: 579–613 (1990).
Selker EU: DNA methylation and chromatin structure: a view from below. Trends Biochem Sci 15: 103–107 (1990).
Smith CJS, Watson CF, Bird CR, Schuch W, Grierson D: Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Mol Gen Genet 224: 477–481 (1990).
Smith CJS, Watson CF, Ray J, Bird CR, Morris PC, Schuch W, Grierson D: Antisense RNA inhibition of polygalacturonase gene expression in transgenic tomatoes. Nature 334: 724–726 (1988).
Tartof KD, Henikoff S: Trans-sensing effects fromDrosophila to humans. Cell 65: 201–203 (1991).
van der Krol AR, Mur LA, Beld M, Mol JNM, Stuitje AR: Flavonoid genes inPetunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2: 291–299 (1990).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Assaad, F.F., Tucker, K.L. & Signer, E.R. Epigenetic repeat-induced gene silencing (RIGS) inArabidopsis . Plant Mol Biol 22, 1067–1085 (1993). https://doi.org/10.1007/BF00028978
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00028978