Abstract
Between 2008 and 2010, 19 strains of infectious bronchitis virus (IBV) were isolated from the vaccinated chicken flocks in Sichuan province, China. The S1 genes of the isolates were amplified and sequenced. Phylogenetic analysis revealed that the 19 isolates and 37 reference IBV strains can be grouped into eight genotypes. Although IBVs of Taiwan-I type, massachusetts type, and proventriculitis type were isolated, but most isolates were LX4 genotype. Homology analysis of the sequences of S1 genes of the 19 isolates and 37 reference IBV strains revealed that the identity of the nucleotides and amino acid sequences of the S1 genes between the 15 LX4-type isolates and other IBV strains were 71.9–99.3% and 72.1–99.1%, respectively, while those of the analyzed IBV of LX4 type were 96.0–99.9% and 94.3–99.8%, respectively. The results from this study and other published results in the GenBank database showed that isolates circulating in Sichuan province in recent years were mainly LX4 genotype, which is the predominant genotype circulated in China in recent years.
Similar content being viewed by others
Introduction
Avian infectious bronchitis (IB) is a common, highly contagious, acute, and economically important viral disease of chickens [1]. The disease affects chickens of all ages, causes respiratory and nephritic syndromes in broilers, and reduces egg production in layers and breeders. The etiologic agent of IB is infectious bronchitis virus (IBV), a member of the family Coronaviridae and genus Coronavirus, replicating primarily in the respiratory tract and also in some epithelial cells of the gut, kidney, and oviduct. Currently, dozens of serotypes and genotypes of IBV have been detected, and many more will be reported in the future. Natural outbreaks of IBV are often the result of infections with strains that differ serologically from the vaccine strains [2, 3], and the antigenicity of IBV was constantly evolving.
IBV is an enveloped, non-segmented, single-stranded, positive-sense RNA virus. Viral genome is around 27.6 kb in length [4, 5] and contains a 5′ cap and a 3′-polyadenylated tail. Its genome encodes four structural proteins: phosphorylated nucleocapsid protein (N), membrane glycoprotein (M), spike glycoprotein(S), and small membrane protein (E) [6, 7]. The S glycoprotein is post-translationally cleaved at protease cleavage recognition motifs into the amino-terminal S1 (92 kDa) and the carboxyl-terminal S2 (84 kDa) subunit by cellular proteases [8]. The protease cleavage motifs are usually associated with one or more pairs of basic amino acids (e.g., Arg-Arg-Phe-Arg-Arg M41) [9]. The multimeric coiled-coil S glycoprotein is extended from the viral membrane, and the S1 glycoprotein is anchored to the viral membrane by the S2 glycoprotein [10].
The S1 glycoprotein carries the neutralizing epitopes and plays an important role in the attachment of the virus to the host cell membrane [9, 11], induction of antibodies for virus neutralization (VN) and hemagglutination-inhibition (HI). In addition, the S1 protein N-terminus plays an important role in tissue tropism and the degree of virulence of the virus. It is well known that the S1 subunit is highly variable; insertions and deletions can be introduced into the viral genome by viral RNA-dependent RNA polymerases which lack proofreading capabilities, and by genetic recombination which occurs via a genomic template-switching mechanism [12]. This has led to the continuous emergence of new IBV serotypes or variants, and has complicated the design of appropriate control programs. Serotypes and genotypes are usually used to character IBV strains. Traditionally, VN and HI tests were used to differentiate and classify IBV strains [13, 14], and monoclonal antibodies have been developed against several serotypes of IBV [15–17], although serotype-specific monoclonal antibodies are only available for a small number of serotypes. In recent researches, S1 gene sequencing and subsequent genetic analysis have provided a fast and accurate method for classifying and predicting IBVs [18–23].
Since the early 1980s, IBV has been diagnosed in China by virus isolation. IB has occurred frequently in flocks and has caused severe economic losses, in spite of the extensive use of vaccines. In China, the available IB vaccines are mainly the Masstype-type strains such as H120, H52, and the Connecticut-type strains such as 28/86 and W93. The serotypes of those vaccines are different from those of the endemic IBV strains, and so bestow only a low degree of protection against the field strains challenge.
In recent years, isolation and sequence analysis of IBV strains in China were frequently reported, and the results showed that there are multiple genotypes of IBV circulated in China [20–23]. However, the nature of IBV strains circulated in Sichuan was not clear, and IBs were epidemic in Sichuan province of China in recent years. The aim of this study was to decipher the natures of the IBV strains circulating in commercial flocks in Sichuan where large number of chickens were raised in recent years. In this article, 19 IBV strains were isolated in Sichuan from clinical outbreaks that occurred in the period of 2008–2010, the whole S1 genes were sequenced, and then sequence alignment and phylogenetic analysis of the isolated IBV strains together with other published IBV strains, and virulence test to 10-day-old chickens were performed. These results provided useful information as to the nature of the IBV strains circulated in China, and the implications of this study in the strategies for future prevention of IBV are also discussed.
Materials and methods
Viruses
The kidney and lung of the diseased chickens were homogenized in phosphate-buffered saline (PBS) containing 200 μ penicillin and 100 μg streptomycin/ml in a ratio of 1:5–10. After 12 h at 4°C, 0.2 ml supernatant was inoculated into the allantoic cavity of 9–11-day-old embryos of specific pathogen-free (SPF) chickens (Beijing experimental animal center, Beijing, China). The embryos were incubated at 37°C and examined twice daily for their viability. The allantoic fluids were harvested from three eggs after 36-h incubation, and three blind passages were conducted until the dwarfing and death of embryos were observed between 48 and 144 h after inoculation. The existence of IBV was verified by reverse transcription-polymerase chain reaction (RT-PCR) of the N protein gene.
Primers for S1 gene
A pair of primers was designed on the basis of the published sequences of spike genes of IBV strains with the aid of computer software Primer Premier 5.0 (PREMIER Biosoft International, Palo Alto, CA, USA); the primers were used to amplify the S1 glycoprotein gene of IBV isolates. Primer sequences were 5′-ACT GAA CAA AAG ACA(C) GAC TTA GT-3′ comprising position 20245-20267 of genomic sequence of IBV strain LX4 and 5′-CCA TAA CTA ACA TAA GGG CAA-3′ comprising position 21946-21966 of genomic sequence of IBV strain LX4. The anticipated amplification segment is about 1.7 kb long encompassing the entire S1 gene. Primers were synthesized by Takara biotechnology (Dalian, China) Co., Ltd.
Viral RNA extraction, RT-PCR amplification, and sequencing
Genomic RNA was extracted from the infected allantoic fluid with Trizol reagent (Takara biotechnology, Dalian, China) according to the manufacturer’s instruction and dissolved in 20 μl of sterile diethyl diethylpyrocarbonate (DEPC)-treated water; the RNA was stored at −70°C until further use. For the reverse transcription (RT) reaction, the Quantscript RT Kit Quant cDNA first strand Synthesis Kit (TIANGEN, Beijing, China) was used, and the total reaction volume was 20 μl, including 2 μl of 10× RT buffer, 2 μl of random 6 mers (10 μM), 2 μl of dNTPmix (2.5 mM each), 1 μl of Quant Reverse Transcriptase, 10 μl of template RNA, and 3 μl Rnase-free water. The reaction mixture was incubated at 37°C for 60 min, and 94°C for 4 min. PCR amplification was carried out using 4 μl cDNA as a template in a total volume of 50 μl containing 25 μl 2× Taq PCR MasterMix (TIANGEN, Beijing, China), 2 μl of 10 μM of each of the two primers, and 17 μl ddH2O. The optimum conditions for PCR were as follows: 94°C for 4 min, 10 cycles at 94°C for 30 s, 55°C for 30 s (a decrease of 0.5°C per cycle), and 72°C for 1 min, and followed by 20 cycles at 94°C for 30 s, 50°C for 30 s, 72°C for 1 min, and a final elongation at 72°C for 10 min. The PCR product was analyzed in 0.9% agarose in Tris–borate–EDTA (TBE) buffer gel containing 0.5 mg/ml ethidium bromide.
DNA cloning
Products of PCR reactions, corresponding to the predicted size of the target gene, were isolated from agarose gels and purified using E.Z.N.A.TM Gel Extraction Kit (Omega, USA). Purified PCR products were ligated with a TA cloning vector pMD18-T (TaKaRa, Japan) and transforming into DH5α Escherichia coli (E. coli) competent cell. Confirmation of clones containing recombinant plasmid was achieved by PCR and restriction enzyme (RE) digestion. The recombinant plasmid was sequenced by Shanghai Sang-gong Biological Engineering Technology & Services Co., Ltd (Shanghai, China).
Sequence analysis of the S1 protein genes
The obtained nucleotide sequences and the deduced amino acid sequences of S1 genes of IBV isolates were aligned using the Editseq program in the Lasergene package (DNASTAR Inc, Madison, WI, USA), and compared with those of 37 other reference IBV strains for the homology analysis with the use of MegAlign program in the same package. Phylogenetic analysis of the nucleotide sequences of the S1 protein gene was performed with the neighbor-joining method using MEGA version 4.0. The bootstrap values were determined from 1000 replicates of the original data. Thirty seven other IBV references strains were chosen for comparison with our isolates, including M41, W93, IBN, 28/86, Ma5, H120, H52, SAIB14, Beaudette, JP8443 (Japan), JP8127 (Japan), 2575/98 (TW I), 2296/95 (TW II), tl/CH/LDT3/03, CK/CH/LSC/95I, SC021202, A2, SAIBK, CK/CH/LSC/99I, DY04, CQ04-1, 4/91, 7/93, Sichuan-06, Spain/05/82, Italy02, J2, Q1, QXIBV, DY05, DY07, CK/CH/LSD/07III, CK/CH/LSD/08I, CK/CH/LJS/08II, CK/CH/LLN/08II, HB08, and LX4. Of these, seven were vaccine strains, nine were isolates from Sichuan province, one was isolate from Chongqing city, six were the unique strains from Japan or Taiwan or the common reference strains such as M41, two were representative isolates associated with proventriculitis, and the rest were mainly the LX4-type strains from other areas of China.
The IBV reference strains were retrieved from the GenBank database, and the backgrounds of the reference strains used in this study and their accession numbers are listed in Table 1.
Virulence test
In order to study the pathogenicity of IBV strains isolated in Sichuan, two representative IBV isolates, CK/CH/SCCD/08I and Sczy3, were used for inoculation in SPF chickens. SPF eggs were obtained from the Beijing Experimental Animal Center (Merial Inc, Beijing, China) and hatched at our lab. SPF chickens were held in separate biosafety level 2 (BSL2) isolators with ad libitum access to food and water and maintained under uniform standard managemental conditions. Experimental and control groups were kept in separate rooms, preventing cross-contamination throughout the course of the experiment. At the age of 10 days, 30 SPF chickens were randomly divided into three groups, each of which consists of 10 chickens. Each chickens in group A and B were inoculated with CK/CH/SCCD/08I and Sczy3, respectively. In each case, the inoculation was done by intranasal route with 100 μl allantoic fluids containing 1 × 105.3 median embryo infectious doses (EID50) of the relevant strain. The control group was mock-inoculated with sterile allantoic fluid. Clinical signs and gross postmortem lesions as well as mortalities were recorded for a period of 30 days after inoculation.
Results
Nineteen IBV strains isolated between 2008 and 2010 in Sichuan province
All the diseased chickens showed typical respiratory symptoms and nephritis, and had not more than 50% mortality. Sixty eight samples were collected from dead or diseased broilers at different chicken flocks located in 10 administrative areas including Yaan, Chengdu, Zigong, Meishan, Mianyang, Leshan, Deyang, Ganzi, Nanchong, and Suining of Sichuan province, China, and 19 IBV strains were isolated. Typical signs, including embryo dwarfing and death, were observed in different passages when each tissue sample or isolate was inoculated into embryos. Diagnosis based on RT-PCR showed all isolates were free of other agents such as Newcastle disease virus and Avian influenza virus. E. coli or Salmonella were isolated in some samples (data not shown). The case histories of local strains are listed in Table 2.
Phylogenetic analysis revealed that LX4 became the predominant genotype of IBV co-circulating in Sichuan area, China
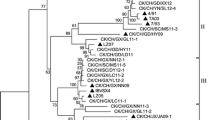
In order to assess the genetic relatedness among the IBV strains, phylogenetic analysis was performed on S1 gene nucleotide sequences consisting of the 19 isolates and 37 other published IBV strains, the 56 IBV strains were separated into eight genetic groups or genotypes (Fig. 1) which were named as LX4, CK/CH/LSC/99I, J2, 4/91, JP, TW, Italy02, and Mass, respectively. Fifteen field isolates from 2008 to 2010, and other nine reference isolates including two Sichuan strains, DY05 and DY07, isolated in 2005 and 2007, respectively, were included in genotype LX4. Eight field reference strains were included in genotype CK/CH/LSC/99I; six strains in this group were isolated from Sichuan and Chongqing between 1995 and 2004. Three IBV strains were included in genotype J2, and isolate CK/CH/SCYA/10I was grouped in this genotype. Three IBV strains were included in genotype 4/91, and Sichuan-06 was 4/91-like strain. Eleven IBV strains were included in genotype Massachusetts, most of which were vaccine strains, and isolate CK/CH/SCNC/08I and CK/CH/SCSN/10I were grouped in this genotype. The selected unique strains from Japan, Taiwan, and Europe formed independent clusters, respectively, in the phylogenetic tree. Remarkably, isolate CK/CH/SCMY/10I was TW-type strain.
Phylogenetic analysis of 19 Sichuan isolates (filled triangle) and 37 reference strains for S1 genes of IBVs (starting at the AUG translation initiation codon and ending at the cleavage recognition motifs). The phylogenetic tree was constructed using the MEGA version 4.0 by the neighbor-joining method with 1000 bootstrap replicates
Low homology of S1 nucleotide and deduced amino acid sequences between IBVs of LX4 type and other genotypes
S1 gene sequences of the 19 IBV isolates were delineated and submitted to the GenBank database (Table 1). Homology analysis based on S1 gene sequences of the 15 LX4-type isolates showed that the homology of the nucleotide and deduced amino acid sequences were 96.0–99.9% and 94.3–99.8%, respectively. However, the homologies of nucleotide and deduced amino acid sequences of the S1 genes between the 15 LX4-type IBV isolates and other IBV strains were 71.9–99.3% and 72.1–99.1%, respectively. The nucleotide and amino acid sequence identity between IBV vaccine strains widely used currently in China such as H120, Ma5, 28/86, 4/91, and the 15 LX4-type isolates were less than 76.7 and 79.0%, respectively. However, those between LX4, a representative strain of the predominant genotype circulated in China in recent years, and the 15 LX4-type isolates were more than 95.4 and 93.5%, respectively. In addition, a phenomenon that deserved attention is that the nucleotide and amino acid sequence identity between IBV strains isolated before 2004 in Sichuan and the 15 LX4-type isolates since 2008 were less than 82.7 and 85.0%, respectively. The nucleotide and amino acid sequence identity between the unique strains from Taiwan or Japan with isolated LX4-type IBV strains were less than 75.1 and 78.5%, respectively. IBV strains of J2 type were originally isolated from proventricus, and the low nucleotide and amino acid sequence identity between IBV strains of J2 genotype and LX4 type were also observed, which were less than 74.1 and 76.1%, respectively.
Alignment analysis of nucleotide and deduced amino acid sequences of S1 protein
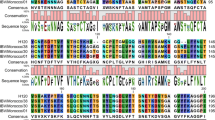
Alignment analysis of S1 complete nucleotide and deduced amino acid sequences of all the 19 isolates and 37 published IBV strains were performed. Nucleotides mutations, insertions, and/or deletions were observed when other IBV strains were compared with IBV H120 strain, resulting in different lengths of nucleotide and amino acids. The S1 genes of the analyzed 56 IBV strains contain 1602, 1611, 1614, 1617, 1620, 1623, 1626, 1629, 1632, and 1635 nucleotides, and 534, 537, 538, 539, 540, 541, 542, 543, 544, and 545 amino acids, respectively (data not shown). The alignment analysis of the deduced amino acid sequences of all the 19 isolates and 21 reference strains revealed that the hypervariable regions (HVRs) were mainly located in three regions, between amino acid residues of 50–79, 115–140, and 275–292 (numbering using S1 sequence of H120 as reference) when compared with H120. The featured deletions, insertions, and mutations are shown in Fig. 2. In addition, two hypervariable sites were also observed, being located at 19–25 and 450–456.
Sequence alignment of S1 amino acid sequences of 40 IBV strains. Dot indicates amino acid identical to that of H120 strain. Dash indicates amino acid deletion in comparison with H120 strain. A alanine, C cysteine, D aspartic acid, E glutamic acid, F phenylalanine, G glycine, H histidine, I isoleucine, K Lysine, L leucine, M methionine, N asparagine, P proline, Q glutamine, R arginine, S serine, T threonine, V valine, Y tyrosine. HVR represent hypervariable region
Virulence study
Clinical symptoms were observed since 3 days’ post challenge, which lasted upto 15 days post challenge in 100% of chickens from both the groups. The chickens showed depression and weakness first, and then became listless and huddled together. 70% (7/10) of the chickens in group A and 80% (8/10) of the chicks in group B suffered from severe gasping, coughing, conjunctivitis, nasal, and ocular discharge. The mortality rates of chickens inoculated by CK/CH/SCCD/08I and Sczy3 were 20% (2/10) and 40% (4/10), respectively. Gross lesions of dead chickens were mainly confined to the kidneys. The kidney parenchyma of the dead birds was pale, swollen and mottled; tubules and urethras were distended with uric acid crystals. The clinical signs of challenged birds tended to disappear gradually after 20 days of inoculation. However, chickens in the control group did not show any symptom and gross lesions observed in the challenged group.
Discussion
Infectious bronchitis (IB) is currently one of the important contagious diseases in poultry production. It not only causes the death of birds, but also causes the chicken to be more sensitive to other pathogens such as E. coli. It can also interfere with the immunization of NDV lentogenic vaccine strain such as Lasota. Immunization failure has frequently occurred and caused severe economic losses in recent years in China, though vaccines such as H120, H52, Ma5, 28/86, and W93 were widely used in flocks. The reason for immune failure is the poor cross-protection between field virus and vaccine strain and the frequent emergence of new variants, and so antigenic characterization of IBV isolates from the recent outbreaks in a region or country is important for selecting the appropriate vaccines and developing new vaccines for the corresponding geographical regions.
IBV strains can be classified into dozens of serotypes by VN test and HI test [3, 14, 24]; subtypes also existed in some serotypes by VN analysis. RT-PCR and sequencing or restriction endonuclease (RE) analysis of S1 gene to characterize IBVs is now increasing [21, 22]. Sequence analysis of field strains has also suggested that the evolution of IBV involves recombination [25–27]. IBV isolates with S gene of very similar sequence can vary substantially in other parts of the genome, and so isolates of the same serotype could belong to different genotypes. Monoclonal antibodies have been developed against several serotype of IBV [15, 28], which can be used to characterize the VN epitopes [17], analyze the antigenic difference of IBVs [16], and cause development of antibody-based blocking of competition ELISA [29].
Since 2008, we have isolated 19 IBV strains from the chicken flocks in different areas of Sichuan province in China. The phylogenetic results showed that the 19 isolates were grouped into four genotypes, but most of the IBV isolates in this study belonged to the LX4 type or QXIBV (15 out of 19). LX4 was isolated by Liu and Kong in 1999 in Xinjiang [18], while QXIBV was isolated by Wang in 1997 in Qingdao, China [30]. In addition, two other IBV strains isolated in Sichuan in 2005 and 2007 were also included in this genotype [20]. However, IBV strains CK/CH/LSC/95I, CK/CH/LSC/99I, SC021202, DY04, and CQ04-1 isolated in Sichuan or Chongqing between 1995 and 2004 were grouped into CK/CH/LSC/99I type. One isolate, CK/CH/SCYA/10I was grouped in J2-type, and two isolates, CK/CH/SCNC/08I and CK/CH/SCSN/10I, were grouped in mass-type, but CK/CH/SCNC/08I was H120-like strain as it showed 99.8% similarity of nucleotide sequence to H120 strain. Remarkably, one IBV strain of TW-type CK/CH/SCMY/10I was isolated, which had not been reported in the mainland of China so far—how did this happen was an enigma. The results from this study and other published results in the GenBank database showed that LX4 genotype has become the prevalent genotype circulated in vaccinated and non-vaccinated flocks in recent years since 2005 in Sichuan province, China. Furthermore, the incidence of LX4- or QX-type IB have been reported in other areas outside China, such as Europe, Africa, and Japan, and the prevalence has been increasing [31–33]. The IBV vaccine strains commercially applied at present in China were correlative mainly with the Massachusetts, such as H120, H52, and 28/86, but it induced poor protection against virulent IBV isolates of different genotypes [34], indicating that vaccines developed from LX4 genotype may be a better choice to control the IBV infections in China.
Alignment analysis results of S1 gene deduced amino acid sequences of all the 19 isolates and 21 reference strains showed that, in addition to the three HVRs reported earlier [20, 22, 35], two other hypervariable sites located at 19–25 and 450–456, respectively, were also observed. Serotype differences of IBVs generally correlated with variations in the HVR of the S1 protein [36, 37], but the significance of HVR in IBV pathogenicity is not clear yet. The unique discontinuous transcription system and the viral polymerase “jumping” possibly contribute to the high RNA recombination frequency in IBVs; Simplot analysis and multiple genes, phylogenetic analysis have been used to detect the recombination events [26].
In conclusion, this study and other published results in the GenBank database have demonstrated that the circulating IBV strains in commercial flocks in Sichuan area in recent years were mainly related to LX4 genotype and were evolving too. The low nucleotides and amino acids identities between the vaccines and the LX4-type isolates may explain why immune failure and outbreak of IB frequently occurred in recent years in commercial flocks. Furthermore, this study suggested the necessity of developing new vaccine strains derived from LX4 genotype. As new genotypes or variant strains may emerge in the future, constant surveillance of new IBV strains is necessary to reveal the molecular character of epidemic IBV strains and to develop better control program of IB in poultry flocks.
References
D. Cavanagha, J. Gelb Jr., Infectious bronchitis, in Disease of Poultry, 12th edn., ed. by Y.M. Saif, A.M. Fadly, J.R. Glisson, L.R. McDougald, L.K. Nolan, D.E. Swayne (Blackwell, Ames, 2008), pp. 117–135
J.K. Cook, Avian Pathol. 13(4), 733–741 (1984). doi:10.1080/03079458408418570
H.N. Wang, Q.Z. Wu, Y. Huang, P. Liu, Avian Dis. 41(2), 279–282 (1997)
M.E. Boursnell, T.D. Brown, I.J. Foulds, P.F. Green, F.M. Tomley, M.M. Binns, J. Gen. Virol. 68(Pt 1), 57–77 (1987)
X.L. Liu, J.L. Su, J.X. Zhao, G.Z. Zhang, Virus Genes. 38(1), 56–65 (2009). doi:10.1007/s11262-008-0282-5
W. Spaan, D. Cavanagh, M.C. Horzinek, J. Gen. Virol. 69(Pt 12), 2939–2952 (1988)
S. Sutou, S. Sato, T. Okabe, M. Nakai, N. Sasaki, Virology. 165(2), 589–595 (1988)
D. Cavanagh, P.J. Davis, D.J. Pappin, M.M. Binns, M.E. Boursnell, T.D. Brown, Virus Res. 4(2), 133–143 (1986). doi:0168-1702(86)90037-7
D. Cavanagh, P.J. Davis, J.K. Cook, D. Li, A. Kant, G. Koch, Avian Pathol. 21(1), 33–43 (1992). doi:10.1080/03079459208418816
D. Cavanagh, Vet. Res. 38(2), 281–297 (2007). doi:10.1051/vetres:2006055
R.L. Parr, E.W. Collissor, Arch. Virol. 133(3–4), 369–383 (1993)
P.S. Masters, Adv. Virus Res. 66, 193–292 (2006). doi:10.1016/S0065-3527(06)66005-3
B.S. Cowen, S.B. Hitchner, Avian Dis. 19(3), 583–595 (1975)
J.K. Cook, A.J. Brown, C.D. Bracewell, Avian Pathol. 16(3), 505–511 (1987). doi:10.1080/03079458708436399
J. Ignjatovic, P.G. McWaters, J. Gen. Virol. 72(Pt 12), 2915–2922 (1991). doi:10.80/03079459708419233
S.A. Naqi, K. Karaca, B. Bauman, Avian Pathol. 22(3), 555–564 (1993). doi:10.1080/03079459308418943
H.G. Niesters, N.M. Bleumink-Pluym, A.D. Osterhaus, M.C. Horzinek, B.A. van der Zeijst, Virology. 161(2), 511–519 (1987). doi:0042-6822(87)90145-0
S.W. Liu, X.G. Kong, Avian Pathol. 33(3), 321–327 (2004). doi:10.1080/0307945042000220697
R. Dolz, J. Pujols, G. Ordonez, R. Porta, N. Majo, Virology. 374(1), 50–59 (2008). doi:10.1016/j.virol.2007.12.020
L.L. Li, C.R. Xue, F. Chen, J.P. Qin, Q.M. Xie, Y.Z. Bi, Y.C. Cao, Vet. Microbiol. 143(2–4), 145–154 (2010). doi:10.1016/j.vetmic.2009.11.022
S.W. Liu, X.N. Zhang, Y. Wang, C. Li, Z. Han et al., Intervirology. 52(4), 223–234 (2009). doi:10.1159/000227134
S.W. Liu, Q.X. Zhang, J.D. Chen, Z.X. Han, X. Liu, L. Feng et al., Arch. Virol. 151(6), 1133–1148 (2006). doi:10.1007/s00705-005-0695-6
C.P. Xu, J.X. Zhao, X.D. Hu, G.Z. Zhang, Vet. Microbiol. 122(1–2), 61–71 (2007). doi:10.1016/j.vetmic.2007.01.006
D.J. King, Avian Dis. 32(2), 335–341 (1988)
J.E. Brooks, A.C. Rainer, R.L. Parr, P. Woolcock, F. Hoerr, E.W. Collisson, Virus Res. 100(2), 191–198 (2004). doi:10.1016/j.virusres.2003.11.016
H.W. Chen, Y.P. Huang, C.H. Wang, Virus Res. 140(1–2), 121–129 (2009). doi:10.1016/j.virusres.2008.11.012
S.A. Kottier, D. Cavanagh, P. Britton, Virology. 213(2), 569–580 (1995). doi:10.1006/viro.1995.0029
P.O. Wainright, P. Villegas, M. Brugh, P.D. Lukert, Avian Dis. 33(3), 482–490 (1989)
K. Karaca, S. Naqi, Vet. Microbiol. 34(3), 249–257 (1993)
Y.D. Wang, Y.L. Wang, Z.C. Zhang, G.C. Fan, Y.H. Jiang, X.E. Liu et al., Chin. J. Anim. Quar. 15(1), 1–3 (1998)
K.J. Worthington, R.J.W. Currie, R.C. Jones, Avian Pathol. 37(3), 247–257 (2008). doi:10.1080/03079450801986529
M.F. Ducatez, A.M. Martin, A.A. Owoade, I.O. Olatoye, B.R. Alkali, I. Maikano et al., J. Gen. Virol. 90(11), 2679–2685 (2009). doi:10.1099/vir.0.012476-0
M. Mase, K. Tsukamoto, K. Imai, S. Yamaguchi, Arch. Virol. 149(10), 1069–2078 (2004). doi:10.1007/s00705-004-0369-9
S.W. Liu, J.F. Chen, Z.X. Han, Q.X. Zhang, Y.B. Shao, X.G. Kong, G.Z. Tong, Avian Pathol. 35(5), 394–399 (2006). doi:10.1080/03079450600920984
R. Smati, A. Silim, C. Guertin, M. Henrichon, M. Marandi, M. Arella et al., Virus Genes. 25(1), 85–93 (2002)
D. Cavanagh, P.J. Davis, A.P. Mockett, Virus Res. 11(2), 141–150 (1988). doi:0168-1702(88)90039-1
S.A. Callison, M.W. Jackwood, D.A. Hilt, Avian Dis. 45(2), 492–499 (2001)
Acknowledgment
This work was financially supported by Program for Changjiang Scholars and Innovative Research Team in University “PCSIRT” (Grant No. IRTO848)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zou, NL., Zhao, FF., Wang, YP. et al. Genetic analysis revealed LX4 genotype strains of avian infectious bronchitis virus became predominant in recent years in Sichuan area, China. Virus Genes 41, 202–209 (2010). https://doi.org/10.1007/s11262-010-0500-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-010-0500-9