Abstract
High-Altitude Andean Lakes (HAAL) of the South American Andes are almost unexplored ecosystems of shallow lakes. The HAAL are recognized by a remarkably high UV exposure, strong changes in temperature and salinity, and a high content of toxic elements, especially arsenic. Being exposed to remarkably extreme conditions, they have been classified as model systems for the study of life on other planets. Particularly, Acinetobacter strains isolated from the HAAL were studied for their survival competence under strong UV-B irradiation. Clinical isolates, Acinetobacter baumannii and Acinetobacter johnsonii, served as reference material. Whereas the reference strains rapidly lost viability under UV-B irradiation, most HAAL-derived strains readily survived this exposure and showed less change in cell number after the treatment. Controls for DNA repair activity, comparing dark repair (DR) or photo repair (PR), gave evidence for the involvement of photolyases in the DNA repair. Comparative measurements by HPLC-mass spectrometry detected the number of photoproducts: bipyrimidine dimers under both PR and DR treatments were more efficiently repaired in the HAAL strains (up to 85 % PR and 38 % DR) than in the controls (31 % PR and zero DR ability). Analysis of cosmid-cloned total genomic DNA from the most effective DNA-photorepair strain (Ver3) yielded a gene (HQ443199) encoding a protein with clear photolyase signatures belonging to class I CPD-photolyases. Despite the relatively low sequence similarity of 41 % between the enzymes from Ver3 and from E. coli (PDB 1DNPA), a model-building approach revealed a high structural homology to the CPD-photolyase of E. coli.
Similar content being viewed by others
Introduction
High-Altitude Andean Lakes (HAAL) of the South American Andes are almost unexplored ecosystems of shallow lakes formed during the Tertiary geological period. They are distributed in the geographical area called the Puna at altitudes between 3,000 and 6,000 m above sea level, and they are isolated from direct human activity (Seufferheld et al. 2008). These lakes suffer from such extreme conditions that they have been classified as model systems for the study of life on other planets (Cabrol et al. 2007).
The HAAL are unique not only for their geographic characteristics and wide range of extreme environmental conditions, but also for their abundant biodiversity (Ordonez et al. 2009). The microbial communities that have evolved within these high-altitude aquatic ecosystems tolerate a wide range of chemical and physical stress such as strong fluctuations in daily temperature, hypersalinity, and variable pH. They have adapted to high levels of UV radiation, a low level of nutrient availability, and high concentrations of heavy metals and metalloids, especially arsenic (Zenoff et al. 2006; Fernandez-Zenoff et al. 2006; Dib et al. 2008, 2009; Ordonez et al. 2009; Farias et al. 2009; Flores et al. 2009). For some of these microorganisms mechanisms have been identified that allow to cope with these extreme conditions, e.g., by the accumulation of polyphosphates (Seufferheld et al. 2008). Hence, the indigenous extremophiles apparently have developed efficient survival strategies and should be able to produce biomolecules adapted to their unusual living conditions that may represent valuable sources of novel bioproducts (e.g. antioxidants, pigments, extremoenzymes) as has been shown for other extremophiles (Sanchez et al. 2009). This topic is intriguing, although, it has not yet been studied in the HAAL ecosystems. The high altitude and low geographical latitude of the HAAL expose these extremophilic communities to high solar irradiance (ca. 165 % higher than at sea level) (Farias et al. 2009). These lakes represent an opportunity to observe the evolution of microorganisms in shallow waters that do not offer substantial UV protection. Survival strategies in these lakes might prove to be ancient and could provide a rare look into Earth’s past. They could also provide critical information for the search for life on other planets (Cabrol et al. 2007).
Previous work identified several strains of Gamma-proteobacteria isolated from the HAAL with a remarkable survival rate, when exposed to high intensity of UV-B irradiation (Zenoff et al. 2006; Fernandez-Zenoff et al. 2006; Ordonez et al. 2009; Flores et al. 2009), most probably an indication for the presence of highly active DNA repair enzymes among other plausible resistance mechanisms.
A great number of specific and highly conserved DNA repair mechanisms has been developed against DNA damage, as there are photoreactivation, excision repair, mismatch repair (MMR), double strand break (DSB) repair. In addition, damage tolerance (dimer bypass), SOS (save our soul) response, checkpoint activation, and programmed cell death (PCD) or apoptosis efficiently act against DNA lesions ensuring the genomic integrity (Sinha and Häder 2002).
Of particular importance is the process of photoreactivation that is executed by photoreactivating enzymes known as “photolyases”, which are well conserved and found throughout the three domains of life. Photolyases are monomeric proteins of 53–66 kDa that contain flavin adenine dinucleotide (FAD) as cofactors and antenna pigments such as deazaflavin or methenyltetrahydrofolate derivatives (Weber 2005). Photolyases are considered among the earliest solutions of nature to the threat of DNA damage by high UV-irradiation intensity (Sancar 2000, 2003). The most common of these lesions is the cis-syn cyclobutane pyrimidine dimer (CPD), formed by a [2 + 2] cycloaddition of two adjacent pyrimidine bases (more frequently a pair of thymines) (Weber 2005). Such CPD lesions bring polymerases to a standstill, eventually leading to cell death. Photolyases bind tightly to CPDs in the dark. Absorption of UV-A and/or blue light (320–470 nm) by the semi-reduced form of their FAD cofactor (FADH•−) starts a downhill electron transfer-driven reaction causing the splitting of the two C–C bonds in the CPD unit and resulting in the re-formation of the two separate pyrimidine bases (Weber 2005).
Among the extremophiles from the HAAL, several strains belonging to the genus Acinetobacter were isolated by us (Ordonez et al. 2009) that are currently subjected to further detailed analysis. The genus Acinetobacter has a long and convoluted taxonomic history dating back to 1911 (Towner 2009). Today, the use of molecular methods has established the identity of 21 different formal species belonging to the genus Acinetobacter (http://www.dsmz.de/microorganisms/bacterial_nomenclature.php). Acinetobacter spp. are widespread and can be isolated from water, soil and living organisms. These gram-negative bacteria are oxidase-negative, non-motile, and strictly aerobic, and tend to be paired cocci (Barbe et al. 2004). Recently, Acinetobacter clinical isolates have received growing attention due to the clinical relevance of their multi-drug resistance (Gordon and Wareham 2010), but less consideration has been given to environmental isolates from Acinetobacter despite of their metabolic versatility, biotechnological potential and their well-known wide resistance to environmental stress (Barbe et al. 2004; Abdel-El-Haleem 2003). The Acinetobacter spp. strains from the HAAL, being isolated from highly UV-B irradiated bacterioplankton and also from a hypersaline environment (Zenoff et al. 2006; Fernandez-Zenoff et al. 2006; Dib et al. 2008; Ordonez et al. 2009), have poly-resistance profiles against UV-B radiation, antibiotics and arsenic, especially Acinetobacter johnsonii A2, a “superbug” isolated from Lake Azul (Fernandez-Zenoff et al. 2006; Dib et al. 2008). Recently, other Acinetobacter spp. strains isolated from Lake Verde and Lake Negra were studied for their high efficient antioxidant enzyme activities in response to UV-B stress (Di Capua et al. 2011).
In order to understand more about the mechanisms involved in the high UV-resistance phenotype displayed by microbes from HAALs isolates belonging to the genus Acinetobacter, the aim of this work was to study the degree of DNA damage and the photo repair potential by repair/survival measurements upon UV-B exposure of four extremophilic Acinetobacter strains from the HAAL ecosystem. In addition, the presence of photolyases has been screened by using molecular methods in combination with 3D modelling, as the observed high survival rate of these microorganisms presumes the presence of novel and efficient repair enzymes.
Materials and Methods
Strains for Study
UV-resistant strains Acinetobacter sp. N40, Ver3, Ver5 and Ver7 from the LIMLA-PROIMI Extremophilic Strain Collection were used (www.limla.com.ar). All the strains had previously been isolated from extreme lakes at the Andean Puna in Argentina (Ordonez et al. 2009). Bacterial strains from DSMZ Bacterial Culture Collection: Acinetobacter johnsonii DSM 6963 and Acinetobacter baumannii DSM 30007 were used as control as its UV-sensitivity as compared to the HAAL strains was previously reported (Di Capua et al. 2011). A. baumannii was also used as a positive control of the PCR-assay for photolyase-coding genes screening, as its full genome sequence reveals a photolyase encoding gene.
Comparison of the phenotypic and genotypic characteristics of Acinetobacter spp. strains isolated from the HAAL and the DSMZ collection strains is given in Table 1.
Phenotypic Characterization
For the morphological assays, the strains were grown for 24 h at 30 °C on LB agar. Gram staining was carried out on 24 h-cultures as described by Doetsch (Doetsch 1981). Cell morphology was visualized using a Nikon microscope. Profiles of enzymatic activities were obtained by using API 20E, API NE20 and API CORYNE test strips (Biomerieux, France) (Albarracin et al. 2010).
Genotypic Characterization
DNA was prepared from cells of the selected strains that were grown on LB broth for 24 h at 30 °C and harvested by centrifugation (3,000 g for 10 min at 4 °C). The pellets were washed twice with distilled water. Total genomic DNA was extracted with the DNeasy Blood and Tissue Kit (Qiagen) following the manufacter’s recommendations.
For taxonomic identification, PCR amplifications were performed in 25 μl reaction volume using universal 16S rRNA gene oligonucleotide primers: 27f and 1492r (Heuer et al. 1997) and ITS (internal transcribed spacers) primers (Daffonchio et al. 1998) in a thermal cycler (Perkin-Elmer, model 9700). PCR products were run in 0.8 % (16S rRNA genes) or 2 % (ITS genes) agarose gel, stained with SYBR Green and visualized using a Gel Doc™ XR + with Image Lab™ software (BioRad). Purification of DNA from gel slabs was performed using QIAquick Gel Extraction Kit (Qiagen), and DNA sequencing was performed by the dideoxy chain termination method with an ABI Prism 3730XL DNA analyzer, using the ABI Prism BigDye terminator cycle sequencing ready reactions kit (PE Biosystems) according to the manufacturer’s protocol. The 16S rRNA sequences reported in this paper have been deposited in GenBank under accession numbers AM778686 (Ver3), AM778688 (Ver5), AM778690 (Ver7) and AM778696 (N40). On the basis of 16s rRNA, Acinetobacter sp. N40 and Ver5 are most closely related to Acinetobacter lwoffii, i.e. 99.8 and 99.9 %, respectively. Acinetobacter sp. Ver3 showed high similarity to Acinetobacter johnsonii (98.8 %) while Ver7 was more closely related with Acinetobacter beijerinckii (98.8 %) (Table 1).
UV-B Resistance Assays and DNA Photo Repair Assays
To evaluate the UV-B resistance and photo repair capability, the selected strains were grown in LB medium at 30 °C with shaking (200 rpm). Cells were harvested in the mid-exponential phase by centrifugation at 8,000 rpm for 30 min at 4 °C. The pellets were washed twice in 0.9 % NaCl and were kept under starvation conditions in the same solution for 12 h at 4 °C. A portion of each cell suspension (OD600: 0.6; 20 ml) was exposed to UV-B irradiation for 10 min (38 kJ m−2) (Vilbert Lourmat VL-4; maximum intensity at 312 nm; average intensity: 7 mW cm−2) with shaking (50 rpm) at 15 °C. Controls were incubated in the dark under the same conditions. After UV-B exposure, 100 μL-aliquots were removed from the tubes and microbial growth was assessed by counting the number of colony-forming units (CFUs) after 48 h of incubation at 30 °C in the dark to prevent photoreactivation.
Both control and UV-B exposed cell suspensions were subjected to photo repair (PR) or dark repair (DR) conditions. Photo repair was allowed by incubating the suspensions under photo-active radiation (PAR) during 120 min (18 W m2) using OSRAM L18 W/77 lamps with continous shaking (50 rpm) at 15 °C, while dark repair was evaluated under the same experimental conditions but in absence of PAR. After each treatment, 100 μL-aliquots were removed from the tubes and the number of CFUs was determined after 48 h of incubation at 30 °C under dark conditions to prevent photoreactivation.
DNA Purification and Bipyrimidine Photoproduct Measurements
For DNA preparation, cell suspensions (10 ml) from different treatments: UV-B exposed for 10 min (UV10), photorepaired cells (PR), dark-repaired cells (DR) and non-exposed controls at initial time (T0), 10 min (T10) and 130 min (T130) of each strain were collected by centrifugation at 3,000 g for 10 min at 4 °C and washed twice with distilled water. Extraction of total genomic DNA was performed in duplicate by using a commercially available genomic DNA kit (DNeasy Blood & Tissue Kit, Qiagen) following the manufacter’s recommendations.
Several types of photoproducts were measured in total genomic DNA from each treatment using previously optimized procedures (Douki et al. 2000; Mouret et al. 2011). Determination was for the four cis-syn CPDs, thymine-thymine (TT), thymine-cytosine (TC), cytosine-thymine (CT), and cytosine-cytosine (CC) sites together with the pyrimidine-(6-4) pyrimidone photoproducts (6-4PPs) at thymine-thymine and thymine-cytosine sites. After extraction, DNA was made soluble into 50 μL of a 0.1 mM deferroxiamine mesylate aqueous solution and then enzymatically hydrolyzed by incubation with nuclease P1, DNase II and phosphodiesterase II (2 h, 37 °C, pH 6), followed by a second digestion step involving phosphodiesterase I and alkaline phosphatase (2 h, 37 °C, pH 8). The resulting solution contained unmodified bases as nucleosides and bipyrimidine photoproducts in the form of dinucleoside monophosphates. All cytosine containing CPDs as well as CT and CC 6-4PPs were analyzed as deaminated products. The digested sample was then injected onto an HPLC system consisting in a Agilent series 1100 system equipped with a Uptisphere ODB reverse phase column (2 × 250 mm ID, particle size 5 μm; Interchim, Montluçon, France). The mobile phase was a gradient of acetonitrile in a 2 mM aqueous solution of triethylammonium acetate. A UV detector set at 260 nm was used to quantify normal nucleosides in order to determine the amount of analyzed DNA. The HPLC flow was then directed toward an API 3000 electrospray triple quadrupole mass spectrometer operating in the negative ionization mode. Under these conditions, the deprotonated pseudomolecular ion of each photoproduct was collected and fragmented in the spectrometer and a specific daughter ion was quantified as previously set-up. External calibration was achieved using authentic reference compounds of varying concentrations, allowing generation of calibration curves. The results were expressed as number of photoproducts per 104 DNA bases.
The assay is very specific and makes possible the individual quantification of each of the different CPDs and 6-4PPs. The technique is sensitive enough to allow the detection of photoproducts in a sample size of few micrograms of DNA extracted from cells or skin (Mouret et al. 2008, 2011).
Cosmid Library Preparation
Cosmid libraries were prepared in the cosmid pWE15 (Stratagene, La Jolla, CA), which has neomycin and ampicillin resistance. Genomic DNA fragments (20–40 kb) obtained after partial Sau3AI digestion of Acinetobacter sp. Ver3 total genomic DNA were ligated into pWE15 linearised by BamHI digestion. Phage packaging and infection of E. coli VCS257 was performed using Giga Pack® III Gold Packaging Extract (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. The transfected E. coli colonies were picked and transferred to 96 well microplates to maintain and store the DNA libraries in DMSO (7 %). Isolation of cosmid DNA from the library was performed using FavorPrep™ 96-Well Plasmid Kit (Favorgen) following the manufacturer’s protocol.
Primer Design and PCR Conditions for Targeting Photolyase Genes
Utilizing the sequence of the photolyase gene from E. coli (phr), a BLAST search was performed in the Acinetobacter sp. strains genome sequences (Acinetobacter sp. ADP1 and Acinetobacter baumannii ATCC 17978). These findings were used to design oligonucleotides for targeting photolyase genes in Acinetobacter strains from HAAL (Table 2).
PCR assays were performed in an automated thermal cycler (Perkin-Elmer, model 9700); annealing temperature conditions were modified until an optimized protocol for each primer was obtained (Table 2). Purification was performed using the illustra GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare), and DNA sequencing on both strands was as described in 2.3. The photolyase coding genes reported in this paper have been deposited in GenBank under accession numbers HQ641332 (Ver7) and HQ443199 (Ver3).
Phylogenetic Analyses
For the phylogenetic analysis of photolyases, the amino acid sequences of the photolyase proteins were aligned using ClustalW2 and the sequences were edited manually using BioEdit (Hall 1999). Maximum likelihood analysis was performed using PhyML3.0 (Guindon and Gascuel 2003), with JTT rate matrix (Jones et al. 1992) and an eight gamma parameter. Bootstrap analysis (100) was performed on most likely tree and the final tree was visualized using iTOL (Letunic and Bork 2007).
Three Dimensional Modeling
Template search was carried out by using the complete sequence from the novel Ver3 photolyase as a query against all three dimensional structures available in RSB PDB online server at www.pdb.org (Berman et al. 2000). The structure that showed the highest score in the analysis (PDB 1DNPA) was used as a template for the modeling performed with Swiss-Pdb Viewer software and Swiss Model server (Guex and Peitsch 1997; Arnold et al. 2006). An energy-minimization program was carried out with the Gromos96 force field implementation of Swiss-PdbViewer (van Gunsteren et al. 1996).
Statistical Analyses
Statistical analyses were conducted using the Microcal™ Origin Working Model Version 6.0. Paired t-test and ONE WAY ANOVA variance analysis were used with a probability level of p < 0.05. All experiments were carried on in duplicates.
Results and Discussion
Survival and Population Recovery of Acinetobacter spp. upon UV-B Exposure
Exposure to UV radiation is considered to be especially harmful to microorganisms due to their small, haploid genomes with little or no functional redundancy, and in general they lack thick, protective cell walls (Ponder et al. 2005). In fact, damage caused by UV in bacterial systems from aquatic environments eventually affects the whole community, having an impact on photosynthesis, biomass production, and the community composition (Winter et al. 2001; Alonso and Pernthaler 2006). Due to the high altitude and the geographic and physicochemical parameters of the HAAL, UV-B radiation is one of the most limiting abiotic factors for bacterioplankton communities (Fernandez-Zenoff et al. 2006; Hernandez et al. 2007). Nevertheless, it was demonstrated that a great diversity of microbes thrive under these harsh conditions (Ordonez et al. 2009). In a previous work, we have demonstrated by a qualitative UV-tolerance test that four Acinetobacter sp. strains isolated from HAAL were more resistant than their closest taxonomical neighbours: A. baumannii DSM 30007, A. johnsonii DSM 6963 and Acinetobacter lwoffii DSM 2403 (Di Capua et al. 2011).
In this work, we present a more quantitative approach to this preliminary finding, i.e., we relate the high UV-tolerance to an efficient DNA-repair system. In this sense, Acinetobacter spp. N40, Ver3, Ver5, Ver7 and the most sensitive strains A. baumannii DSM 30007 and A. johnsonii DSM 6963 were exposed to UV-B radiation for 10 min (38 kJ m−2) in saline solution and then subjected to photo and dark repair assays.
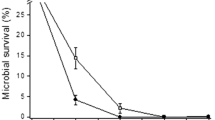
The control strains were not able to maintain their population after the UV-B exposure, while Acinetobacter sp. Ver7 showed practically 100 % survival after the irradiation, Acinetobacter sp. Ver5, 50 %, and Acinetobacter sp. Ver3 and N40, 20 % and 1 %, respectively (Fig. 1). These results are similar to the qualitative findings by Di Capua et al. (2011) who showed that Acinetobacter sp. Ver7 was the more resistant strain together with Ver3 when exposed to 9 kJ m−2 whereas Acinetobacter sp. Ver5 and N40 were less resistant. These slight differences in behaviour could be explained by the different conditions tested in our work, i.e. starvation during 12 h before exposure, pre-incubation of strains before plating and higher UV-B dose. Nevertheless, in both cases, we found a marked inhibition of survival of the control strains upon UV-B exposure supporting the fact that HAAL representatives are more resistant than closely related clinical strains.
Survival of Acinetobacter spp. strains after UV-B exposure for 10 min (38 kJ m−2), photo and dark repair assays. T 0 . Initial population. T 10 , Non-exposed control after 10 min. T 130 , Non-exposed control after 130 min. UV 10 , Resulting population after 10 min of UV-B exposure. DR, UV-B exposed population allowed to dark repair during 120 min. PR, UV-B exposed population allowed to photo repair during 120 min. The clinical isolates A. baumannii and A. johnsonii were used as sensitive controls
After irradiation-induced DNA damage, the strains were immediately exposed to conditions conducive to photo repair (PR) and dark repair (DR) during a period of 120 min, respectively. Interestingly, Acinetobacter sp. Ver3 and Ver5 were able to completely recover their initial population after PR (Fig. 1). Acinetobacter sp. N40 was also able to increase its population thanks to the PR (from 1 % to 60 %). In contrast, A. baumannii was not able to recover after PR, while A. johnsonii recovered partially by 15 %.
DR was less efficient for recovering of the initial population for all strains (Fig. 1). A. baumannii and A. johnsonii were not able to recover after DR, while Acinetobacter sp. Ver3, Ver5 and N40 recovered partially to 21, 27, and 30 %, respectively.
UV-B resistance among the HAAL’s Acinetobacter seems to be the rule; apart from the strains described in this work, we have also reported on Acinetobacter strains able to resist up to 12–24 h of continous UV-B irradiation (Ordonez et al. 2009; Flores et al. 2009). These reports are the only ones of Acinetobacter spp. strains being isolated from strongly UV-B exposed bacterioplankton (mean value of UV-B irradiance reached 10.8 W m−2 for the 300- to 325-nm range).
Total DNA Bypirimidine Photoproduct Accumulation upon UV-B Exposure and Their Relation with Survival of the Selected Strains
In parallel to survival/recovery experiments, also the amount of DNA photoproducts was determined after each treatment using HPLC-ESI-MS/MS measurements including the four types of CPDs and the two main 6-4PPs in total genomic DNA of selected strains. This method has been previously designed for the study of UV-induced DNA damage in skin, as well as several eukaryotic and prokaryotic cells. It is based on fully defined reference compounds and has become a routine method for the quantitative determination of photoproducts (Douki et al. 2000; Mouret et al. 2008, 2011).
Although HAAL strains displayed higher UV-B resistance profiles, they also showed the highest number of photoproducts after 10 min of UV-B irradiation (Fig. 2), roughly 25 % more photoproducts than the controls. This may suggest that UV-B resistance ability is not strictly related to a lower DNA photodamage. This pattern of high survival plus high photoproducts accumulation could alternatively be explained by a DNA lesion bypass, a typical mechanism of the error-prone dark repair that keeps the cell active with DNA replication (by polymerase V) without DNA damage repair (Tanooka et al. 1991; Smith and Walker 1998). This was recently demonstrated for Acinetobacter johnsonii A2, a strain isolated from Lake Azul at the HAAL that accumulated high content of CPDs in its DNA after UV-B damage (Fernandez-Zenoff et al. 2006).
Number of total photoproducts produced in N40: Acinetobacter sp. N40; Ver3: Acinetobacter sp. Ver3; Ver5: Acinetobacter sp. Ver5; Ver7: Acinetobacter sp. Ver7;. B: Acinetobacter baumannii. J: Acinetobacter johnsonii after UV-B exposure (38 kJ m-2) for 10 min (UV 10 ) and photo (PR) and dark repair (DR) treatments. Lesions are expressed per 104 base-pairs of the DNA extracted from the cell material after each treatment. The number of lesions in the non-irradiated controls are not shown as they are, in all cases, below 0.5 lesion per 104 bases. The clinical isolates A. baumannii and A. johnsonii were used as sensitive controls
Acinetobacter spp. Ver5, N40, Ver3 and Ver7 displayed 8.8, 8.4, 7.3, and 6.1 lesions per 104 bases in their DNA. Acinetobacter sp. Ver7 accumulated lower numbers of photoproducts than the other strains and showed 100 % survival, while Acinetobacter sp. N40 presented the highest photoproduct accumulation (Fig. 2). Accordingly, almost the entire initial population of N40 was depleted after the UV-B exposure taking in account the CFU numbers (Fig. 1). Nevertheless, Acinetobacter sp. Ver5 which exhibited 5 % more photoproducts than N40 (Fig. 2) showed only a 50 % decrease of its initial population (Fig. 1). It is evident that DNA photodamage itself is not the only factor that determines cell viability, as UV-B irradiation also alters survival by direct or indirect effects on lipids and proteins (Buma et al. 2003). Aquatic organisms have a variety of physiological and biochemical mechanisms available for reducing the damage incurred by exposure to UV, including screening, quenching and repair (Banaszak 2003). Thus, it is not unexpected that each strain from the HAAL may display different mechanisms of photoprotection for vital cellular components, i.e., UV-absorbing compounds, antioxidants and carotenoids. In all Acinetobacter strains tested, photoprotection mechanisms due to pigments may be excluded, as no HAAL strain showed any pigmentation. Recently, an unusually high catalase activity was found in Acinetobacter sp. Ver3 and Ver7 as well as good a tolerance to pro-oxidants (H2O2 and methylviologen) (Di Capua et al. 2011), indicating that also an efficient antioxidant machinery can explain the high UV-resistance fitness in these two bacteria. In fact, inhibition of the catalase activity led to a decrease in UV-B tolerance for strain Ver7 (Di Capua et al. 2011).
The control strains, A. johnsonii and A. baumannii accumulated only 6.5 and 6.1 lesions per 104 bases (Fig. 2), a value similar to that measured in strain Ver7, although this extent of DNA damage did not allow them to maintain their population after UV-B exposure (Fig. 1). In this case, the target of UV-B damage could be other than DNA (lipids or proteins), and/or these strains were unable to cope with even a very low DNA damage. This low survival level combined with low number of CPD formation in the sensitive strains have also been reported for Serratia marcescens MF42 and the marine Pseudomonas putida strain 2IDINH (Fernandez-Zenoff et al. 2006).
Photo- and Dark Repair Abilities upon UV-B Exposure of the Selected Strains
The most efficient strains that were found to deplete DNA bypyrimidine photoproducts under both PR and DR treatments were Acinetobacter sp. Ver3 and Ver7 with a reduction of 69 and 85 % (PR) and 22 and 38 % (DR), respectively (Fig. 2). To a lower extent, Acinetobacter sp. N40 was able to reduce the level of photoproducts under PR (54 %) and DR (25 %). Among the HAAL strains, Acinetobacter sp. Ver5 was the least efficient one to repair DNA lesions as it reduced them only under PR (27 %). Nevertheless, it is interesting to note that irrespective of the number of initial DNA lesions, all HAAL strains recovered their population totally or to a significant extent under PR (Fig. 1). In contrast, A. baumannii did not display significant photo repairing or dark repairing capacities (Fig. 2) whereas A. johnsonii has a behavior similar to Ver5: under photorepair, it achieved 31 % of reduction of DNA lesions whereas dark repair was not observed. Even if the photorepair capability of A. johnsonii was similar to strain Ver5 (Fig. 2), it would not allow recovering of its population to the same extent as strain Ver5 (Fig. 1). This finding supports the idea that the HAAL strains can cope with a higher percentage of DNA damage, even for accumulated mutations in great quantities. This trait may be a distinctive feature for UV-resistant strains. The high mutagenicity predisposition of HAAL strains was previously observed in antibiotic-resistant strains (Dib et al. 2008), and concurs with recently found abundant repetitive sequences in a HAAL megaplasmid (Wagenknecht et al. 2010), being another strong argument that these microbes are used to deal with strong DNA damage. One might also keep in mind that strong mutation rates can be advantageous for adaptation and survival in highly UV-exposed environments with strongly changing environmental conditions.
The distribution of photoproducts and the relative yield of 6-4PPs and CPDs were also evaluated (Figs. 3 and 4). The DNA photoproduct distribution does not show recognizable trends with the exception that A. baumannii exhibited a larger proportion of TT damage than the others strains tested (Fig. 3), although the GC content is similar for all. Because TT-CPD has been the first identified and was found to be the most frequent DNA photoproduct in mammalian cells, it was hypothesized that DNA with lower thymine content would be less sensitive to UV radiation (Singer and Ames 1970). However, this hypothesis was immediately challenged (Bak et al. 1972), and in fact, recent studies have reported a poor correlation between UV resistance in bacterial species and their GC content (Matallana-Surget et al. 2008), a finding that concurs well with our results.
Yields of CPD-photoproducts as number of lesions per 104 base-pairs in the DNA extracted from the selected strains after the different treatments. T 0 , Initial population. T 10 , Non-exposed control after 10 min. T 130 , Non-exposed control after 130 min. UV 10 , Remaining population after 10 min of UV-B exposure. DR, UV-B exposed population allowed to darkrepair during 120 min. PR, UV-B exposed population allowed to photo repair during 120 min. N40: Acinetobacter sp. N40; Ver3: Acinetobacter sp. Ver3; Ver5: Acinetobacter sp. Ver5; Ver7: Acinetobacter sp. Ver7;. B: Acinetobacter baumannii. J: Acinetobacter johnsonii
Yields of 6,4-photoproducts expressed as the number of lesions per 104 pairs of nucleotides in the DNA extracted from the selected strains after the different treatments. T 0 , Initial population. T 10 , Non-exposed control after 10 min. T 130 , Non-exposed control after 130 min. UV 10 , Resulting population after 10 min of UV-B exposure. DR, UV-B exposed population allowed to dark repair during 120 min. PR, UV-B exposed population allowed to photo repair during 120 min. N40: Acinetobacter sp. N40; Ver3: Acinetobacter sp. Ver3; Ver5: Acinetobacter sp. Ver5; Ver7: Acinetobacter sp. Ver7;. B: Acinetobacter baumannii. J: Acinetobacter johnsonii
On the average, the total yield of CPDs is more important (Fig. 3) than the sum of TT and TC 6-4 PPs (Fig. 4) with a ratio varying between 3.5 (N40) and 3.1 (Ver5). In most cases, the TT cyclobutane pyrimidine dimer is the predominant photoproduct with a decreasing order for the other CPDs: TT>TC>CT>CC (Fig. 4). It may also be noted that the frequency of TC 6-4PP which is about 5- to 6-fold higher than that of TT 6-4PP (Fig. 4) is within the range of the formation yield of TC-CPD (Fig. 4).
Considering both, the photoreaction and dark excision repairs of the bipyrimidine photoproducts, it is likely that the latter process is taking place during the photolyase-mediated reversion of CPDs. This may suggest -as the incubation period of PR and DR treatments is the same- that the reported yield of photoreversion of CPDs is overestimated by the contribution of nucleotide excision repair. As a first general observation, it was found that the photoreversion efficacy of bipyrimidine photoproducts that varies according to the investigated strains, concerns only CPDs (Fig. 3), in agreement with previous findings in several microorganisms (Gascon et al. 1995; Joux et al. 1999; Agogue et al. 2005; Fernandez-Zenoff et al. 2006; Matallana-Surget et al. 2008). PR of CPDs was found to be highly efficient in our extremophiles, mainly strain Ver7 and, to a lesser extent, strain Ver3. The photo repair of CPDs was lower in Ver5, N40 and A. johnsonii while almost inexistent in A. baumannii. Significant differences were also noted for dark repair of bipyrimidine photoproducts. For a given strain, the excision of the main TT and TC CPDs and 6-4PPs was similar. This is in contrast with the preferential removal of 6-4 PPs in eukaryotic cells (Courdavault et al. 2005). The most efficient excision of both CPDs and 6-4PPs is observed for strain Ver7 by comparison with that of Ver3 and Ver5 strains. On the other hand, no significant DR is noted for Ver5, A. baumannii and A. johnsoni (Figs. 3 and 4); these differences in PR and DR efficiencies provide a good support for the observed differences in sensitivities of the investigated strains to UV-B radiation with the exception of the Ver5 microorganism which is quite photoresistant despite a lack of DR and a relatively low PR efficiency.
Screening for Photolyase Genes in Acinetobacter spp. Strains
The high resistance found for these strains and the just discussed active photo repair implies the presence of highly efficient repair systems for UV-B induced DNA damage. Bacteria have several repair mechanisms in response to such damages, i.e., dark repair and photoreactivation. Photoreactivation, being light-dependent, employs a photolyase enzyme through activation by different wavelengths, such as UV-A (315 to 340 nm) and photosynthetic active radiation (PAR) (400 to 700 nm). In bacteria, this repair activity is limited to CPDs, as confirmed in our results where the repair of 6-4PPs is not better under PR than DR (Fig. 4).
The potential presence of photolyase-encoding genes in the most resistant strains was investigated by PCR screening, using Acinetobacter-specific degenerate primers from highly conserved regions of photolyases (Table 2). From this approach, unambiguously positive amplification products (ca. 250 bp) were obtained for two of the tested strains: Acinetobacter sp. Ver3 and Ver7. A BLASTX analysis revealed for both of these sequences a high similarity to the conserved region of photolyases. The partial sequence from strain Ver3 showed 94 % similarity to the photolyase from Acinetobacter sp. ATCC 27244 (ZP_03822952). Similarly, the partial sequence from strain Ver7 (HQ641332) showed a strong similarity (95 %) to the photolyase from Acinetobacter sp. ATCC 27244. This high degree of similarity is clearly due to the highly conserved sequence motifs of photolyases that were used for PCR primer design. Next, a cosmid DNA library (1,000 clones) was constructed from Ver3 allowing a search for the full-length gene of the putative photolyase. A PCR-based screening yielded one clone for which the formerly identified photolyase sequence was confirmed. This cosmid clone was directly sequenced by primer walking, yielding an ORF (1,413 bp, Accession No. HQ443199) encoding a protein with signatures typical for a DNA photolyase. The translated sequence was BLASTed against the non-redundant protein sequences deposited in the NCBI database. The nearest match in BLAST analysis was the photolyase from Acinetobacter johnsonii SH046 (ZP_06062937) which showed 60 % identity revealing the sequence novelty of the Ver3 photolyase protein.
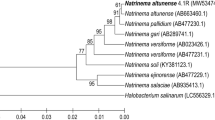
The maximum likelihood phylogenetic tree constructed from the nearest match obtained during BLAST analysis revealed that the photolyases from Ver3 and Ver7 are grouped together in a subfamily (Fig. 5). These putative photolyases are placed between the photolyase proteins from Acinetobacter sp. ADP1 environmental strain and other pathogenic Acinetobacter spp. Arrangement of these novel sequences with representative photolyases in the phylogenetic tree clearly documents their kinship with photolyases, however, identifies them also as relatively distant and novel members of this protein family.
Maximum-likelihood tree constructed from putative photolyase sequences from Ver3 (Accession no. HQ443199) and Ver7 (Accession no. HQ641332) along with selected photolyases from various gamma proteobacteria from NCBI database (these two entries 2nd and 3rd from top are identified by arrows). The tree is midpoint rooted and the solid circles on the nodes indicate the bootstrap values >70. (Abbreviations: A. : Acinetobacter, P.: Pseudomonas)
It is obvious from the phylogenetic analysis that the sequences from the two environmental strains are more related to each other than any of them is related to the pathogenic A. baumannii strains (Fig. 5). This indicates that environmental bacteria may harbor quite different photolyases than the pathogenic ones. The original living conditions of the Ver strains in Laguna Verde (4,400 m asl), located in volcanic settings at the Andean region of Catamarca province, Argentina, (27° 38’S, 65° 32’W), are challenging, as metabolic enzymatic functions normally occurring within the cells need to be adapted to these extremely harsh conditions. As the lakes are shallow and these bacteria were collected from the surface water, they suffer from a high UV-B irradiance reaching up to 10.8 W m−2 for the 300 to 325 nm range (Flores et al. 2009). The conditions prevailing in the water are not less challenging; the arsenic content is 0.8 mg L−1 (the allowed As content in drinking water is 0.01 mg L−1) while the salinity is 5 % (sea salinity is around 3.5 %). The phosphorous and chlorophyll contents are low (<12 and 1.04 μg/L, respectively) indicating the oligotrophic nature of this lake (Flores et al. 2009). It can thus be assumed that the herein described photolyases exhibit novel functional properties (considering the hypersalinity and high contents of arsenic) in accordance with their extreme origin, as may be documented from further biochemical analysis of the gene product functions.
Structural Modelling of Photolyase from Acinetobacter sp. Ver3 and Preliminary Functional Evidence
Based on sequence alignments and secondary structure predictions for the novel photolyase from the HAAL and entries in the PDB data bank, we found the highest three- dimensional similarity to the photolyase from E. coli (PDB 1DNPA). This photolyase belongs to the class I CPD-photolyases (Müller and Carell 2009), and was the first for which a three dimensional structure was determined (Park et al. 1995). In this group, there are also other crystallized CPD-photolyases from the cyanobacteria Anacystis nidulans (Tamada et al. 1997), the eubacterium Thermus thermophilus (Klar et al. 2006) and the archaeon Sulfolobus tokodaii (Fujihashi et al. 2007). Even as the sequence similarity of Ver3 with that of E. coli is low (41 %), we found a high kinship between the 3D structures as could be demonstrated from a model-building approach that indicated a nearly congruent protein folding (Fig. 6a). The structure revealed a proximal alpha-beta domain and a distal helical domain that binds to FAD in full accordance to the structure of the E. coli photolyase (Park et al. 1995). The N-terminal anti-parallel bundle of beta sheets enclosed by alpha helices is a typical folding motif of photolyases. The residues Asp105, Glu106, Lys298, Glu368 and Leu380 that interact with the antenna chromophore 5,10-methenyltetrahydrofolylpolyglutamate (MTFH) are fully conserved between the E. coli enzyme and the Ver3 photolyase, while in place of cys292 (E. coli protein) there is a serine residue (Ser297) in Ver3 photolyase. The residues Tyr225, Thr237, Ser238, Leu240, Ser241, Trp276, Arg283, Trp343, Asn346, Asp377, Asp379, and Ala382 that interact with FAD are all conserved in Ver3 except Gln239 that replaces an arginine residue (Arg236) in E. coli photolyase. The chain of three tryptophan residues (Trp311, Trp364, and Trp387) participating in electron transfer reaction (Brettel and Byrdin 2010) is also conserved in Ver3 photolyase. The structural arrangement of this Trp-triade demonstrates the close proximity of these three residues to the isoalloxazine ring of FAD (Fig. 6b). This arrangement supports the putative CPD-photolyase property of this protein which agrees with the efficient ability of Ver3 for repairing CPDs lesions.
a Structural model of Ver3 photolyase using the crystal structure of E. coli photolyase (PDB 1DNPA) as a template. The MTHF cofactor is seen on the top right of the structure and the FAD cofactor is embedded in the middle. b Structural arrangement of FAD and three tryptophan residues (that take part in electron transfer) in the 3D model structure from Ver3 photolyase
Despite a high level of sequence diversity, the topological similarity between all photolyase structures known today is one of the most surprising discoveries. Müller and Carrell (Müller and Carell 2009) calculated the total root mean square deviation for 9 structures (including photolyases and cryptochromes) with varying sequences and found only 1.914 Å over 369 residues, as calculated with Secondary-Structure Matching (SSM) (Krissinel and Henrick 2004) (http://www.ebi.ac.uk/msd-srv/ssm). It seems that all these proteins utilize similar energy and electron transfer steps which, owing to the sensitivity of these processes, require a precise setting of distance and orientation changes. This apparently forces the proteins to maintain a common fold in order to ensure an optimal arrangement of the essential cofactors (Epple and Carell 1999).
A more detailed in vitro analysis of this novel photolyase is under way; yet, preliminary observations proof a functional enzyme: E. coli cells (BL21) that were transformed with the gene putatively encoding the photolyase have been exposed to UV-B irradiation (38 kJ m−2) during cell growth. Of the control cells (untransformed E. coli), more than 99 % did not survive this treatment, whereas 48 % of the cells expressing the recombinant photolyase did survive the same dose of UV-B exposure (Pathak et al., unpublished data). Further biochemical characterization of this novel photolyase is currently performed.
Concluding Remarks
In this work, we add knowledge to the biology of extremophiles belonging to the genus Acinetobacter that were isolated from the HAAL. Due to the outstandingly harsh conditions to which microorganisms from HAAL are exposed, their survival strategies provide helpful information for astrobiological research. Not by chance, the HAAL region had been classified as a model area for the living conditions on the early earth that might have been similar to other planets in the universe. The here described photolyase-based, remarkably strong repair activity in the studied Acinetobacter strain, the adaptation to “lethal” arsenic concentrations, and the accumulation of repetitive sequences in their DNA point to the various survival strategies as a consequence to the aggressive environmental conditions in HAAL. In this study, we have quantitatively determined the UV-B resistance patterns of four extremophilic Acinetobacter strains isolated from the HAAL ecosystem and we have demonstrated that this UV-resistance phenotype is related to an efficient machinery of photoreactivation involving, at least, a CPD-photolyase, but it also shows the capability of the strains to cope with high content of photoproducts in their DNA. We have determined the sequences of photolyase-coding genes (one complete and one partial) in two of the environmental isolates. Up to now, the only photolyase sequence information was obtained from genomic projects of some Acinetobacter strains. Here we demonstrate for the first time the existence of novel photolyases belonging to extremophilic Acinetobacter strains.
We also show that this Ver3 photolyase displays low sequence similarity to the class I CPD-photolyase of E. coli (41 %). Yet, an overlay of both structures reveals a high kinship between both 3D geometries. This arrangement supports the putative CPD-photolyase activity of the Ver3 protein which agrees with the efficient capability of the strain for repairing CPDs lesions. Its heterologous expression will allow studies on its photochemical properties, will “shine light” on its potential outstanding properties and will reveal the molecular basis of its enzymatic function.
References
Abdel-El-Haleem D (2003) Acinetobacter: environmental and biotechnological applications. Afr J Biotechnol 2:71–74
Agogue H, Joux F, Obernosterer I, Lebaron P (2005) Resistance of marine bacterioneuston to solar radiation. Appl Environ Microbiol 71:5282–5289
Albarracin VH, Alonso-Vega P, Trujijjo ME, Amoroso MJ, Abate CM (2010) Amycolatopsis tucumanensis sp nov., a copper-resistant actinobacterium isolated from polluted sediments. Int J Syst Evol Microbiol 60:397–401
Alonso C, Pernthaler J (2006) Roseobacter and SAR11 dominate microbial glucose uptake in coastal North Sea waters. Environ Microbiol 8:2022–2030
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Bak AL, Atkins JF, Meyer SA (1972) Evolution of DNA base compositions in microorganisms. Science 175:1391–1393
Banaszak AT (2003) Photoprotective physiological and biochemical response of aquatic organisms to UVR. In: Helbling EW, Zagarese H (eds) UV effects in aquatic organisms and ecosystems. The Royal Society of Chemistry, Cambridge, pp 329–356
Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, Labarre L et al (2004) Unique features revealed by the genome sequence of Acinetobacter sp ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res 32:5766–5779
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H et al (2000) The protein data bank. Nucleic Acids Res 28:235–242
Brettel K, Byrdin M (2010) Reaction mechanisms of DNA photolyase. Curr Opin Struct Biol 20:693–701
Buma AG, Boelen P, Jeffrey WH, Helbling E, Zagarese H (2003) UVR-induced DNA damage in aquatic organisms. In: Helbling EW, Zagarese H (eds) UV effects in aquatic organisms and ecosystems. Royal Society of Chemistry, Cambridge, pp 291–327
Cabrol NA, Grin EA, Kiss KT, Ács E, Grigorszky I, Szabò K, Tóth B, Fike DA, Hock AN, Demergasso C, Escudero L, Chong G, Galleguillos P, Grigsby BH, Zambrana Román J, McKay CP, Tambley C (2007) Signatures of habitats and life in earth’s high-altitude lakes: clues to Noachian aqueous environments on mars. In: Chapman M (ed) The geology of mars. Cambridge University Press, pp. 349–370
Courdavault S, Baudouin C, Charveron M, Canguilhem B, Favier A, Cadet J, Douki T (2005) Repair of the three main types of bipyrimidine DNA photoproducts in human keratinocytes exposed to UVB and UVA radiations. DNA Repair 4:836–844
Daffonchio D, Borin S, Frova G, Manachini PL, Sorlini C (1998) PCR fingerprinting of whole genomes: the spacers between the 16S and 23S rRNA genes and of intergenic tRNA gene regions reveal a different intraspecific genomic variability of Bacillus cereus and Bacillus licheniformis. Int J Syst Bacteriol 48:107–116
Di Capua C, Bortolotti A, Farias ME, Cortez N (2011) UV-resistant Acinetobacter sp. solates from Andean wetlands display high catalase activity. FEMS Microbiol Lett 317:181–189
Dib J, Motok J, Zenoff VF, Ordonez O, Farias ME (2008) Occurrence of resistance to antibiotics, UV-B, and arsenic in bacteria isolated from extreme environments in high-altitude (above 4400 m) andean wetlands. Curr Microbiol 56:510–517
Dib JR, Weiss A, Neumann A, Ordoñez O, Estévez MC, Farías ME (2009) Isolation of bacteria from remote high altitude Andean lakes able to grow in the presence of antibiotics. Recent Patents Antiinfect Drug Discov 4:66–76
Doetsch RN (1981) Determinative methods of light microscopy. In: Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GB (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington, pp 21–33
Douki T, Court M, Sauvaigo S, Odin F, Cadet J (2000) Formation of the main UV-induced thymine dimeric lesions within isolated and cellular DNA as measured by high performance liquid chromatography-tandem mass spectrometry. J Biol Chem 275:11678–11685
Epple R, Carell T (1999) Efficient light-dependent DNA repair requires a large cofactor separation. J Am Chem Soc 121:7318–7329
Farias ME, Fernandez-Zenoff V, Flores R, Ordonez O, Estevez C (2009) Impact of solar radiation on bacterioplankton in Laguna Vilama, a hypersaline Andean lake (4650 m). J Geophys Res 114:G00D04. doi:10.1029/2008JG000784
Fernandez-Zenoff V, Sineriz F, Farias ME (2006) Diverse responses to UV-B radiation and repair mechanisms of bacteria isolated from high-altitude aquatic environments. Appl Environ Microbiol 72:7857–7863
Flores MR, Ordoñez OF, Farías ME (2009) Isolation of UV-B resistant bacteria from two high altitude Andean lakes (4,400 m) with saline and non saline conditions. J Gen Appl Microbiol 55:447–458
Fujihashi M, Numoto N, Kobayashi Y, Mizushima A, Tsujimura M, Nakamura A et al (2007) Crystal structure of archaeal photolyase from sulfolobus tokodaii with two FAD molecules: implication of a novel light-harvesting cofactor. J Mol Biol 365:903–910
Gascon J, Oubina A, Perez Lezaun A, Urmeneta J (1995) Sensitivity of selected bacterial species to Uv-radiation. Curr Microbiol 30:177–182
Gordon NC, Wareham DW (2010) Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int J Antimicrob Agents 35:219–226
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hernandez KL, Quinones RA, Daneri G, Farias ME, Helbling EW (2007) Solar UV radiation modulates daily production and DNA damage of marine bacterioplankton from a productive upwelling zone (36°S), Chile. J Exp Mar Biol Ecol 343:82–95
Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH (1997) Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol 63:3233–3241
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Joux F, Jeffrey WH, Lebaron P, Mitchell DL (1999) Marine bacterial isolates display diverse responses to UV-B radiation. Appl Environ Microbiol 65:3820–3827
Klar T, Kaiser G, Hennecke U, Carell T, Batschauer A, Essen LO (2006) Natural and non-natural antenna chromophores in the DNA photolyase from Thermus thermophilus. ChemBioChem 7:1798–1806
Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D: Biol Crystallogr 60:2256–2268
Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128
Matallana-Surget S, Meador JA, Joux F, Douki T (2008) Effect of the GC content of DNA on the distribution of UVB-induced bipyrimidine photoproducts. Photochem Photobiol Sci 7:794–801
Mouret S, Charveron M, Favier A, Cadet J, Douki T (2008) Differential repair of UVB-induced cyclobutane pyrimidine dimers in cultured human skin cells and whole human skin. DNA Repair 7:704–712
Mouret S, Bogdanowicz P, Haure M-J, Castex-Rizzi N, Cadet J, Favier A, Douki T (2011) Assessment of the photoprotection properties of sunscreens by chromatographic measurement of DNA damage in skin explants. Photochem Photobiol 87:109–116
Müller M, Carell T (2009) Structural biology of DNA photolyases and cryptochromes. Curr Opin Struct Biol 19:277–285
Ordonez OF, Flores MR, Dib JR, Paz A, Farias ME (2009) Extremophile culture collection from Andean lakes: extreme pristine environments that host a wide diversity of microorganisms with tolerance to UV radiation. Microb Ecol 58:461–473
Park HW, Kim ST, Sancar A, Deisenhofer J (1995) Crystal-structure of DNA photolyase from Escherichia-coli. Science 268:1866–1872
Ponder MA, Gilmour SJ, Bergholz PW, Mindock CA, Hollingsworth R, Thomashow MF, Tiedje JM (2005) Characterization of potential stress responses in ancient Siberian permafrost psychroactive bacteria. FEMS Microbiol Ecol 53:103–115
Sancar GB (2000) Enzymatic photoreactivation: 50 years and counting. Mutat Res Fundam Mol Mech Mutagen 451:25–37
Sancar A (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev 103:2203–2237
Sanchez LA, Gomez FF, Delgado OD (2009) Cold-adapted microorganisms as a source of new antimicrobials. Extremophiles 13:111–120
Seufferheld MJ, Alvarez HM, Farias ME (2008) Role of polyphosphates in microbial adaptation to extreme environments. Appl Environ Microbiol 74:5867–5874
Singer CE, Ames BN (1970) Sunlight ultraviolet and bacterial dna base ratios. Science 170:822–826
Sinha RP, Häder D-P (2002) UV-induced DNA damage and repair: a review. Photochem Photobiol Sci 1:225–236
Smith BT, Walker GC (1998) Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics 148:1599–1610
Tamada T, Kitadokoro K, Higuchi Y, Inaka K, Yasui A, deRuiter PE et al (1997) Crystal structure of DNA photolyase from Anacystis nidulans. Nat Struct Biol 4:887–891
Tanooka H, Tanaka K, Shinozaki K (1991) Heterospecific expression of misrepair-enhancing activity of mucab in Escherichia coli and Bacillus subtilis. J Bacteriol 173:2906–2914
Towner KJ (2009) Acinetobacter: an old friend, but a new enemy. J Hosp Infect 73:355–363
van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE et al (1996) Biomolecular simulation: the GROMOS96 manual and user guide. Vdf Hochschulverlag AG an der ETH Zürich, Zürich
Wagenknecht M, Dib JR, Thürmer A, Farias ME, Meinhardt M (2010) Structural peculiarities of linear megaplasmid, pLMA1, from Micrococcus luteus interfere with pyrosequencing reads assembly. Biotechnol Lett 32:1853–1862
Weber S (2005) Light-driven enzymatic catalysis of DNA repair: a review of recent biophysical studies on photolyase. Biochim Biophys Acta 1707:1–23
Winter C, Moeseneder MM, Herndl GJ (2001) Impact of UV radiation on bacterioplankton community composition. Appl Environ Microbiol 67:665–672
Zenoff V, Heredia J, Ferrero M, Sineriz F, Farias ME (2006) Diverse UV-B resistance of culturable bacterial community from high-altitude wetland water. Curr Microbiol 52:359–362
Acknowledgments
The authors acknowledge the generous financial support by the PICT-MPI 2006 01090 Project (FONCyT, Argentina) the Proalar Agreement (MINCYT-DAAD) DA/09/05 and MPI Bioinorganic Chemistry, Germany. V.H.A. was supported by DAAD and Marie Curie (FP7-UE) scholarships. M.E.F., C.D.B. and V.H.A. are researchers from the National Research Council (CONICET) in Argentina. The authors gratefully acknowledge helpful discussions with Prof. Dr. Silvia Braslavsky.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Albarracín, V.H., Pathak, G.P., Douki, T. et al. Extremophilic Acinetobacter Strains from High-Altitude Lakes in Argentinean Puna: Remarkable UV-B Resistance and Efficient DNA Damage Repair. Orig Life Evol Biosph 42, 201–221 (2012). https://doi.org/10.1007/s11084-012-9276-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-012-9276-3