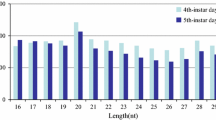
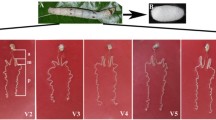
Abstract
MicroRNAs (miRNAs) are a class of non-protein coding small RNAs that regulate expression of genes at post-transcriptional levels. Increasing evidence has shown that miRNAs play multiple roles in biological processes, including development, cell proliferation and apoptosis. Based on the conservation of miRNAs sequence, using a computational homology search based on genomic survey sequence analysis, a total of 16 novel miRNAs were identified and characteristics such as family and evolutionary conservation have been described. By using these newly identified miRNAs, the mRNA database of silkworm was blasted and 21 potential targets of miRNAs were detected. Most of these miRNA targeted genes were predicted to encode transcription factors. The semi-stem loop RT–PCR based assays were performed and found that silkworm miRNAs have diverse expression patterns during development.



Similar content being viewed by others
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Hannon GJ (2002) RNA interference. Nature 418:244–251
Avnit-Sagi T, Kantorovich L, Kredo-Russo S, Hornstein E, Walker MD (2009) The promoter of the pri-miR-375 gene directs expression selectively to the endocrine pancreas. PLoS One 4:e5033
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901–906
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294:853–858
Berezikov E, Cuppen E, Plasterk RH (2006) Approaches to microRNA discovery. Nat Genet 38:S2–S7
Griffiths-Jones S (2004) The microRNA registry. Nucleic Acids Res 32:D109–D111
Griffiths-Jones S (2006) miRBase: the microRNA sequence database. Methods Mol Biol 342:129–138
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158
Wang XJ, Reyes JL, Chua NH, Gaasterland T (2004) Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol 5:R65
Tong CZ, Jin YF, Zhang YZ (2006) Computational prediction of microRNA genes in silkworm genome. J Zhejiang Univ Sci B 7:806–816
Liu S, Xia Q, Zhao P, Cheng T, Hong K, Xiang Z (2007) Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori). BMC Dev Biol 7:88
He PA, Nie Z, Chen J, Lv Z, Sheng Q, Zhou S, Gao X, Kong L, Wu X, Jin Y, Zhang Y (2008) Identification and characteristics of microRNAs from Bombyx mori. BMC Genom 9:248
Yu X, Zhou Q, Li SC, Luo Q, Cai Y, Lin WC, Chen H, Yang Y, Hu S, Yu J (2008) The silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PLoS One 3:e2997
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115:787–798
Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005) Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res 33:e179
Zhou ZS, Huang SQ, Yang ZM (2008) Bioinformatic identification and expression analysis of new microRNAs from Medicago truncatula. Biochem Biophys Res Commun 374:538–542
Cao J, Tong C, Wu X, Lv J, Yang Z, Jin Y (2008) Identification of conserved microRNAs in Bombyx mori (silkworm) and regulation of fibroin L chain production by microRNAs in heterologous system. Insect Biochem Mol Biol 38:1066–1071
Zhang Y, Zhou X, Ge X, Jiang J, Li M, Jia S, Yang X, Kan Y, Miao X, Zhao G, Li F, Huang Y (2009) Insect-specific microRNA involved in the development of the silkworm Bombyx mori. PLoS One 4:e4677
Zeng Y (2006) Principles of micro-RNA production and maturation. Oncogene 25:6156–6162
Guddeti S, Zhang DC, Li AL, Leseberg CH, Kang H, Li XG, Zhai WX, Johns MA, Mao L (2005) Molecular evolution of the rice miR395 gene family. Cell Res 15:631–638
Chang DT, Wang CC, Chen JW (2008) Using a kernel density estimation based classifier to predict species-specific microRNA precursors. BMC Bioinform 9(12):S2
Xu JH, Li F, Sun QF (2008) Identification of microRNA precursors with support vector machine and string kernel. Genom Proteom Bioinform 6:121–128
Zhang B, Pan X, Stellwag EJ (2008) Identification of soybean microRNAs and their targets. Planta 229:161–182
Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI (2008) The birth and death of microRNA genes in Drosophila. Nat Genet 40:351–355
Zhang R, Peng Y, Wang W, Su B (2007) Rapid evolution of an X-linked microRNA cluster in primates. Genome Res 17:612–617
Niwa R, Slack FJ (2007) The evolution of animal microRNA function. Curr Opin Genet Dev 17:145–150
Iwasaki H, Takahashi M, Niimi T, Yamashita O, Yaginuma T (1997) Cloning of cDNAs encoding Bombyx homologues of Cdc2 and Cdc2-related kinase from eggs. Insect Mol Biol 6:131–141
Champlin DT, Truman JW (1998) Ecdysteroid control of cell proliferation during optic lobe neurogenesis in the moth Manduca sexta. Development 125:269–277
Yoshida I, Moto K, Sakurai S, Iwami M (1998) A novel member of the bombyxin gene family: structure and expression of bombyxin G1 gene, an insulin-related peptide gene of the silkmoth Bombyx mori. Dev Genes Evol 208:407–410
Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC (2005) The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev 19:2947–2952
Acknowledgments
We gratefully acknowledge the technique assistance of Dr. Y. X. Yu at Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing, China. This research was partly supported by research grants form the National High Tech Development Project of China, the 863 Program (2007AA100504) and the National Basic Research 973 Program (2005CB121004).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, Y., Zou, Q., Tang, S.M. et al. Computational identification and characteristics of novel microRNAs from the silkworm (Bombyx mori L.). Mol Biol Rep 37, 3171–3176 (2010). https://doi.org/10.1007/s11033-009-9897-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-009-9897-4