Abstract
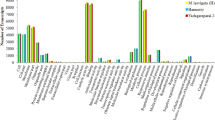
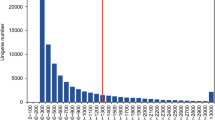
Mulberry is an important economic crop spread widely in China for long time, and cutting is the most suitable method for good varieties in large scale propagation, which is limited by the low adventitious rooting rate. Currently, the regulation mechanisms in adventitious roots (AR) formation of mulberry (Morus alba L.) are not completely known. A genome-wide gene expression profiling was conducted by Solexa sequencing at pivotal stages. Differentially expressed annotated genes were obtained from the analysis of Gene Orthology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Orthology (KO). Some related genes were selected to confirm the altered expression levels by quantitative real-time PCR (qRT-PCR). By transcript profiling analysis we found differentially expressed genes associated with common pathways including hormone signal transduction, cell cycle, carbohydrate metabolism and energy metabolism that play important roles in AR formation (M. alba L), which were found more obvious in the period of root primordia formation, not initial formation of callus. In addition, the qRT-PCR confirmations agreed well with the results. The period of root primordia formation was verified to be more important during the whole procedure of AR formation in cuttings, mainly regulated by several critical signal pathways of hormones and effected by other genes in cell cycle and energy metabolism.

Similar content being viewed by others
References
Adams TS, Eissenstat DM (2014) The continuous incorporation of carbon into existing Sassafras albidum fine roots and its implications for estimating root turnover. PLoS One 9:e95321. doi:10.1371/journal.pone.0095321
Ahkami AH, Lischewski S, Haensch KT, Porfirova S, Hofmann J, Rolletschek H, Melzer M et al (2009) Molecular physiology of adventitious root formation in Petunia hybrida cuttings: involvement of wound response and primary metabolism. New Phytol 181:613–625
Ansorge WJ (2009) Next-generation DNA sequencing techniques. New Biotechnol 25:195–203
Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7:986–995
Bashline L, Li S, Gu Y (2014) The trafficking of the cellulose synthase complex in higher plants. Ann Bot 114:1059–1067. doi:10.1093/aob/mcu040
Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M et al (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14:3073–3088
Brinker M, van Zyl L, Liu W, Craig D, Sederoff RR, Clapham DH, von Arnold S (2004) Microarray analyses of gene expression during adventitious root development in Pinus contorta. Plant Physiol 135:1526–1539
Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71:684–697
Chen Y, Hao X, Cao J (2014) Small auxin upregulated RNA (SAUR) gene family in maize: identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and Sorghum. J Integr Plant Biol 56:133–150. doi:10.1111/jipb.12127
De Klerk G-J, van der Krieken W, de Jong J (1999) Review the formation of adventitious roots: new concepts, new possibilities. In Vitro Cell Dev Biol Plant 35:189–199
Du Wei, Cheng Jia-Ling (2014) Morphological and Anatomical observation on cortex rooting process of Mulberry (Morus L.) Greenwood cuttings. Acta Sericologica Sin 40:13–17 (in Chinese)
Feng S et al (2015) Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J Integr Plant Biol 57:783–795. doi:10.1111/jipb.12327
Gao J, Zeng XF, Liu XH, Yang SX (2011) Cutting propagation of Periploca forrestii and dynamic analyses of physiological and biochemical characteristics related to adventitious roots formation. Zhong yao cai = Zhongyaocai = J Chin Med Mater 34:841–845
Geiss G, Gutierrez L, Bellini C (2009) Adventitious root formation: new insights and perspectives, annual plant reviews, vol 37: root development. Wiley, New York, pp 127–156
Gibson SI (2005) Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol 8:93–102
Greenwood MS, Cui X, Xu F (2001) Response to auxin changes during maturation-related loss of adventitious rooting competence in loblolly pine (Pinus taeda) stem cuttings. Physiol Plant 111:373–380
Gutierrez L, Mongelard G, Flokova K, Pacurar DI, Novak O, Staswick P, Kowalczyk M et al (2012) Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 24:2515–2527
Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49:373–385
Hermans C, Porco S, Verbruggen N, Bush DR (2010) Chitinase-like protein CTL1 plays a role in altering root system architecture in response to multiple environmental conditions. Plant Physiol 152:904–917. doi:10.1104/pp.109.149849
Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D et al (2004) Transcript profiling of early lateral root initiation. Proc Natl Acad Sci USA 101:5146–5151
Iliev EA, Xu W, Polisensky DH, Oh MH, Torisky RS, Clouse SD, Braam J (2002) Transcriptional and posttranscriptional regulation of Arabidopsis TCH4 expression by diverse stimuli. Roles of cis regions and brassinosteroids. Plant Physiol 130:770–783. doi:10.1104/pp.008680
Ivanchenko MG, Muday GK, Dubrovsky JG (2008) Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J 55:335–347
Jain M, Tyagi AK, Khurana JP (2006) Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88:360–371
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T et al (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Kant S, Bi YM, Zhu T, Rothstein SJ (2009) SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol 151:691–701
Kazan K, Manners JM (2012) MYC2: the Master in Action. Mol Plant 6:686–703. doi:10.1093/mp/sss128
Kell DB (2012) Large-scale sequestration of atmospheric carbon via plant roots in natural and agricultural ecosystems: Why and How. Philos Trans R Soc Lond B Biol Sci 367:1589–1597. doi:10.1098/rstb.2011.0244
Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110:213–222
Li C, Schilmiller AL, Liu G, Lee GI, Jayanty S, Sageman C, Vrebalov J et al (2005) Role of beta-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 17:971–986
Li YJ, Fu YR, Huang JG, Wu CA, Zheng CC (2011) Transcript profiling during the early development of the maize brace root via Solexa sequencing. FEBS J 278:156–166
Li T, Qi X, Zeng Q, Xiang Z, He N (2014) MorusDB: a resource for mulberry genomics and genome biology. Database 2014. doi:10.1093/database/bau054
Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49:387–400
Lisewski AM et al (2014) Supergenomic network compression and the discovery of EXP1 as a glutathione transferase inhibited by artesunate. Cell 158:916–928. doi:10.1016/j.cell.2014.07.011
Liu P et al (2013) Transcript profiling of microRNAs during the early development of the maize brace root via Solexa sequencing. Genomics 101:149–156. doi:10.1016/j.ygeno.2012.11.004
Majer C, Xu C, Berendzen KW, Hochholdinger F (2012) Molecular interactions of rootless concerning crown and seminal roots, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.). Philos Trans R Soc Lond B Biol Sci 367:1542–1551
Marchant A, Bhalerao R, Casimiro I, Eklof J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14:589–597
Mau CH (2010) The progress of sericulture during the yuan period (13th–14th century) studies on Nongsang Jiyao. Rev Synth 131:193–217
Meldau S, Baldwin IT, Wu J (2011) For security and stability: SGT1 in plant defense and development. Plant Signal Behav 6:1479–1482
Mishra BS, Singh M, Aggrawal P, Laxmi A (2009) Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS One 4:e4502
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Pan G, Lou C (2008) Isolation of an 1-aminocyclopropane-1-carboxylate oxidase gene from mulberry (Morus alba L.) and analysis of the function of this gene in plant development and stresses response. J Plant Physiol 165:1204–1213. doi:10.1016/j.jplph.2007.02.012
Peret B, Larrieu A, Bennett MJ (2009) Lateral root emergence: a difficult birth. J Exp Bot 60:3637–3643
Proudnikov D, Yuferov V, Zhou Y, LaForge KS, Ho A, Kreek MJ (2003) Optimizing primer–probe design for fluorescent PCR. J Neurosci Methods 123:31–45
Raya-Gonzalez J, Pelagio-Flores R, Lopez-Bucio J (2012) The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana. J Plant Physiol 169:1348–1358
Sebastian J et al (2015) PHABULOSA controls the quiescent center-independent root meristem activities in Arabidopsis thaliana. PLoS Genet 11:e1004973. doi:10.1371/journal.pgen.1004973
Sorin C, Negroni L, Balliau T, Corti H, Jacquemot MP, Davanture M, Sandberg G et al (2006) Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol 140:349–364
Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inze D, Peer WA et al (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70:978–990
Takahashi F, Sato-Nara K, Kobayashi K, Suzuki M, Suzuki H (2003) Sugar-induced adventitious roots in Arabidopsis seedlings. J Plant Res 116:83–91
Wagner GP, Kin K, Lynch VJ (2012) Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci = Theorie in den Biowissenschaften 131:281–285. doi:10.1007/s12064-012-0162-3
Wan X, Landhausser SM, Lieffers VJ, Zwiazek JJ (2006) Signals controlling root suckering and adventitious shoot formation in aspen (Populus tremuloides). Tree Physiol 26:681–687
Wang C, Yu H, Zhang Z, Yu L, Xu X, Hong Z, Luo L (2015) Phytosulfokine is involved in positive regulation of Lotus japonicus nodulation. Mol Plant Microbe Interact 28:847–855. doi:10.1094/mpmi-02-15-0032-r
Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7:1555–1567
Zheng J, Xiao X, Zhang Q, Yu M, Xu J, Wang Z (2015) Maternal protein restriction induces early-onset glucose intolerance and alters hepatic genes expression in the peroxisome proliferator-activated receptor pathway in offspring. J Diabetes Investig 6:269–279
Acknowledgments
This work was supported by China Agriculture Research System (Grant No. CARS-22).
Author contribution list
Guarantor of integrity of entire study: Jialing Cheng, Xiaolong Du, Xiaofeng Zhang. Study concepts: Jialing Cheng, Xiaolong Du, Xiaofeng Zhang. Study design: Xiaolong Du, Xiaofeng Zhang. Literature research: Xiaolong Du, Xiaofeng Zhang, Hao Nie. Experimental studies: Xiaolong Du, Xiaofeng Zhang, Minglu Liu. Data acquisition: Xiaolong Du, Xiaofeng Zhang, Minglu Liu, Hao Nie. Data analysis/interpretation: Xiaolong Du, Xiaofeng Zhang. Manuscript preparation: Xiaolong Du, Xiaofeng Zhang. Manuscript definition of intellectual content: Jialing Cheng, Xiaolong Du, Xiaofeng Zhang. Manuscript editing: Xiaolong Du, Xiaofeng Zhang. Manuscript revision/review: Jialing Cheng, Xiaolong Du, Xiaofeng Zhang. Manuscript final version approval: Jialing Cheng.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
XiaoLong Du and XiaoFeng Zhang have contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Du, X., Zhang, X., Nie, H. et al. Transcript profiling analysis reveals crucial genes regulating main metabolism during adventitious root formation in cuttings of Morus alba L. . Plant Growth Regul 79, 251–262 (2016). https://doi.org/10.1007/s10725-015-0130-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-015-0130-2