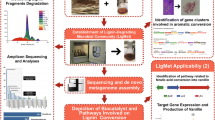
Abstract
A number of prokaryotes actively contribute to lignin degradation in nature and their activity could be of interest for many applications including the production of biogas/biofuel from lignocellulosic biomass and biopulping. This review compares the reliability and efficiency of the culture-dependent screening methods currently used for the isolation of ligninolytic prokaryotes. Isolated prokaryotes exhibiting lignin-degrading potential are presented according to their phylogenetic groups. With the development of bioinformatics, culture-independent techniques are emerging that allow larger-scale data mining for ligninolytic prokaryotic functions but today, these techniques still have some limits. In this work, two phylogenetic affiliations of isolated prokaryotes exhibiting ligninolytic potential and laccase-encoding prokaryotes were determined on the basis of 16S rDNA sequences, providing a comparative view of results obtained by the two types of screening techniques. The combination of laboratory culture and bioinformatics approaches is a promising way to explore lignin-degrading prokaryotes.


Similar content being viewed by others
References
Ahmad M, Taylor CR, Pink D, Burton K, Eastwood D, Bendingb GD, Timothy DH (2010) Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders. Mol BioSyst 6:815–821
Ahmad M, Roberts JN, Hardiman EM, Singh R, Eltis LD, Bugg TD (2011) Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 50:5096–5107
Andrade C, Pereira N, Antranikian G (1999) Extremely thermophilic microorganisms and their polymer-hydrolytic enzymes. Rev Microbiol 30:287–298
Ausec L, Zakrzewski M, Goesmann A (2011) Bioinformatic analysis reveals high diversity of bacterial genes for laccase-like enzymes. PLoS One 6:1–9
Banat IM, Nigam P, Singh D, Marchant R (1996) Microbial decolorization of textile-dye containing effluents: a review. Bioresour Technol 58:217–227
Bandounas L, Wierckx NJP, de Winde JH, Ruijssenaars HJ (2011) Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol 11:94
Benner R, Maccubbin AE, Hodson RE (1984a) Preparation, characterization, and microbial degradation of specifically radiolabeled [14C]lignocelluloses from marine and freshwater macrophytes. Appl Environ Microbiol 47:381–389
Benner R, Newell SY, Maccubbin AE, Hodson RE (1984b) Relative contributions of bacteria and fungi to rates of degradation of lignocellulosic detritus in salt-marsh sediments. Appl Environ Microbiol 48:36–40
Björdal CG, Nilsson T, Daniel G (1999) Microbial decay of waterlogged archaeological wood found in Sweden applicable to archaeology and conservation. Int Biodeterior Biodegrad 43:63–73
Björkman A (1956) Studies on finely divided wood. Part I. Extraction of lignin with neutral solvents. Svensk Papperstidning 59:477–485
Boer W, Folman LB, Summerbell RC, Boddy L (2005) Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29:795–811
Boudet A-M (1998) A new view of lignification. Trends Plant Sci 3:67–71
Bourbonnais R, Paice MG (1990) Oxidation of non-phenolic substrates: an expanded role for laccase in lignin biodegradation. FEBS Lett 267:99–102
Brown ME, Chang MC (2014) Exploring bacterial lignin degradation. Curr Opin Chem Biol 19:1–7
Browning BL (1967) Methods of wood chemistry. Interscience, New York
Brune A, Ohkuma M (2011) Role of the termite gut microbiota in symbiotic digestion. In: Biology of termites: a modern synthesis. Springer Netherlands, Dordrecht, pp 439–475
Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R (2011a) Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 28:1883–1895
Bugg TDH, Ahmad M, Hardiman EM, Singh R (2011b) The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol 22:394–400
Chandra R, Bharagava RN (2013) Bacterial degradation of synthetic and kraft lignin by axenic and mixed culture and their metabolic products. J Environ Biol 34:991–999
Chandra R, Singh S (2009) Isolation and characterization of bacterial strains Paenibacillus sp. and Bacillus sp. for kraft lignin decolorization from pulp paper mill waste. J Gen Appl Microbiol 54:399–407
Chen CY, Huang YC (2013) Properties of the newly isolated extracellular thermo-alkali-stable laccase from thermophilic actinomycetes, Thermobifida fusca and its application in dye intermediates oxidation. AMB Express 3:1–9
Childers SE, Ciufo S, Lovley DR (2002) Geobacter metallireducens accesses insoluble Fe (III) oxide by chemotaxis. Nature 416:767–769
Colodette JL, Zikeli F, José F, Gomes B, Rio JC (2013) Detailed characterization of black liquor (lignin) deriving from kraft and soda-antraquinone pulping. Paper presented at the 8th International Black Liquor Colloquium. Federal University of Minas Gerais, Belo Horizonte
Colombini MP, Lucejko JJ, Modugno F, Orlandi M, Tolppa E-L, Zoia L (2009) A multi-analytical study of degradation of lignin in archaeological waterlogged wood. Talanta 80:61–70
Copley SD, Rokicki J, Turner P, Daligault H, Nolan M, Land M (2012) The whole genome sequence of Sphingobium chlorophenolicum L-1: insights into the evolution of the pentachlorophenol degradation pathway. Genome Biol Evol 4:184–198
Corvini PFX, Schäffer A, Schlosser D (2006) Microbial degradation of nonylphenol and other alkylphenols—our evolving view. Appl Microbiol Biotechnol 72:223–243
Cowling EB (1961) Comparative biochemistry of the decay of sweetgum sapwood by white-rot and brown-rot fungi. US Dept. of Agriculture, Washington
Crawford DL, Crawford RL (1976) Microbial degradation of lignocellulose: the lignin component. Appl Environ Microbiol 31:714–717
Dai X, Zhu Y, Luo Y, Song L, Liu D et al (2012) Metagenomic insights into the fibrolytic microbiome in yak rumen. PLoS One 7(7):e40430. doi:10.1371/journal.pone.0040430
Davis JR, Sello JK (2010) Regulation of genes in Streptomyces bacteria required for catabolism of lignin-derived aromatic compounds. Appl Microbiol Biotechnol 86:921–929
DeAngelis KM, Gladden JM, Allgaier M (2010) Strategies for enhancing the effectiveness of metagenomic-based enzyme discovery in lignocellulolytic microbial communities. Bioenergy Res 3:146–158
DeAngelis KM, Allgaier M, Chavarria Y, Fortney JL, Hugenholtz P, Simmons B, Sublette K, Silver WL, Hazen TC (2011) Characterization of trapped lignin-degrading microbes in tropical forest soil. PLoS One 6(4):e19306. doi:10.1371/journal.pone.0019306
DeAngelis KM, Sharma P, Varney R, Simmons B, Isern N, Markilllie YM, Nicora C, Norbeck AD, Taylor RC, Aldrich JT, Robinson EW (2013) Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1. Front Microbiol 4:1–13
Deng Y, Fong SS (2011) Metabolic engineering of Thermobifida fusca for direct aerobic bioconversion of untreated lignocellulosic biomass to 1-propanol. Metab Eng 13:570–577
Deschamps AM, Mahoudeau G, Conti M, Lebeault JM (1980a) Bacteria degrading tannic acid and related compounds. J Ferment Technol 58:93–97
Deschamps AM, Mahoudeau G, Lebeault JM (1980b) Fast degradation of kraft lignin by bacteria. Appl Microbiol Biotechnol 9:45–51
Deschamps AM, Gillie JP, Lebeault JM (1981) Direct delignification of untreated bark chips with mixed cultures of bacteria. Appl Microbiol Biotechnol 13:222–225
Dhindwal S, Patil DN (2011) Biochemical studies and ligand-bound structures of biphenyl dehydrogenase from Pandoraea pnomenusa strain B-356 reveal a basis for broad specificity of the enzyme. J Biol Chem 286:37011–37022
Dhouib A, Hamza M, Zouari H, Mechichi T, H’midi R, Labat M, Martínez MJ, Sayadi A (2005) Autochthonous fungal strains with high ligninolytic activities from Tunisian biotopes. Afr J Biotechnol 4:431–436
Diaz E, Ferrandez A, Prieto MA, Garcia JL (2001) Biodegradation of aromatic compounds by Escherichia coli. Microbiol Mol Biol Rev 65:523–569
Diaz E, Jiménez JL, Nogales J (2012) Aerobic degradation of aromatic compounds. Environ Biotechnol 24:431–442
Duan RB (2008) A microbial catalyst preservations and preparation method. China Patent
Eggeling L, Sahm H (1980) Degradation of coniferyl alcohol and other lignin-related aromatic compounds by Nocardia sp. DSM 1069. Arch Microbiol 126:141–148
Emerson D, Chauhan S, Oriel P, Breznak JA (1994) Haloferax sp. D1227, a halophilic Archaeon capable of growth on aromatic compounds. Arch Microbiol 161:445–452
Faison BD, Kirk TK (1983) Relationship between lignin degradation and production of reduced oxygen species by Phanerochaete chrysosporium. Appl Environ Microbiol 46:1140–1145
Geszvain K, McCarthy JK, Tebo BM (2013) Elimination of Manganese(II, III) oxidation in Pseudomonas putida GB-1 by a double knockout of two putative multicopper oxidase Genes. Appl Environ Microbiol 79:1357–1366
Girault R, Peu P, Béline F, Lendormi T, Guillaume S (2013) Caractéristiques des substrats et interactions dans les filières de co-digestion: cas particulier des co-substrats d'origine agro-industrielle. Sci Eaux Territ 3:44–53
González JM, Whitman WB, Hodson RE, Moran MA (1996) Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl Environ Microbiol 62:4433–4440
González J, Mayer F, Moran M, Hodson R, Whitman W (1997) Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Bacteriol 47:369–376
Gonzalez JM, Mayer F, Moran MA, Hodson RE, Whitman WB (1997) Sagittula stellata gen. nov., sp. nov., a lignin-transforming bacterium from a coastal environment. Int J Syst Bacteriol 47:773–780
Graf N, Altenbuchner J (2014) Genetic engineering of Pseudomonas putida KT2440 for rapid and high-yield production of vanillin from ferulic acid. Appl Microbiol Biotechnol 98(1):137–149
Grbic-Galic D, Pat-Polasko LL (1985) Enterobacter cloacae DG-6: a strain that transforms methoxylated aromatics under aerobic and anaerobic conditions. Curr Microbiol 12:321–324
Guermazi S, Daegelen P, Dauga C, Rivière D, Bouchez T, Godon JJ, Gyapay G, Sghir A, Pelletier E, Weissenbach J (2008) Discovery and characterization of a new bacterial candidate division by an anaerobic sludge digester metagenomic approach. Environ Microbiol 10:2111–2123
Haider K (1966) Synthese von 14C-ringmarkierten phenolischen ligninspaltstficken und ligninalkoholen aus Ba14CO3. J Label Compd 2:174–183
Haider K, Trojanowski J (1975) Decomposition of specifically 14C-labelled phenols and dehydropolymers of coniferyl alcohol as models for lignin degradation by soft and white rot fungi. Arch Microbiol 105:33–41
Haider K, Trojanowski J, Sundman V (1978) Screening for lignin degrading bacteria by means of 14C-labelled lignins. Arch Microbiol 119:103–106
Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5:R245–R249
Haritash A, Kaushik C (2009) Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater 169:1–15
Heo S, Kwak J (2006) Characterization of an extracellular xylanase in Paenibacillus sp. HY-8 isolated from an herbivorous longicorn beetle. J Microbiol Biotechnol 16:1753–1759
Hervé V, Le Roux X, Uroz S, Gelhaye E, Frey-Klett P (2014) Diversity and structure of bacterial communities associated with Phanerochaete chrysosporium during wood decay. Environ Microbiol 16:2238–2252
Hess M, Sczyrba A, Egan R, Kim T-W, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467
Heym B, Alzari PM, Honore N, Cole ST (2006) Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol 15:235–245
Hongoh Y, Deevong P (2005) Intra-and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl Environ Microbiol 71:6590–6599
Huang XF, Santhanam N, Badri DV, Hunter WJ, Manter DK, Decker SR, Vivanco JM, Reardon KF (2013) Isolation and characterization of lignin-degrading bacteria from rainforest soils. Biotechnol Bioeng 110:1616–1626
Jadhav JP, Phugare SS, Dhanve RS, Jadhav SB (2010) Rapid biodegradation and decolorization of Direct Orange 39 (Orange TGLL) by an isolated bacterium Pseudomonas aeruginosa strain BCH. Biodegradation 21:453–463
Janshekar H, Fiechter A (1982) On the bacterial degradation of lignin European. J Appl Microbiol Biotechnol 14:47–50
Jokela J, Pellinen J, Salkinoja-Salonen M, Brunow G (1985) Biodegradation of two tetrameric lignin model compounds by a mixed bacterial culture. Appl Microbiol Biotechnol 23:38–46
Kalyani DC, Phugare SS, Shedbalkar UU, Jadhav JP (2011) Purification and characterization of a bacterial peroxidase from the isolated strain Pseudomonas sp. SUK1 and its application for textile dye decolorization. Ann Microbiol 61:483–491
Kato K, Kozaki S, Sakuranaga M (1998) Degradation of lignin compounds by bacteria from termite guts. Biotechnol Lett 20:459–462
Kerr TJ, Kerr RD (1987) Microorganism having characteristics of an Arthrobacter capable of degrading peanut hull lignin. U.S. Patent No. 4,643,899, 17 Feb 1987
Kerr TJ, Kerr RD, Benner R (1983) Isolation of a Bacterium capable of degrading peanut hull lignin. Appl Environ Microbiol 46:1201–1206
Kirby R (2006) Actinomycetes and lignin degradation. Adv Appl Microbiol 58:125–168
Kirk TK, Connors WJ, Bleam RD, Hackett WF, Zeikus JG (1975) Preparation and microbial decomposition of synthetic [14C]lignins (lignin biodegradation/wood decay/dehydrogenative polymerizate). Proc Natl Acad Sci U S A 72:2515–2519
Klason P (1910) Determining of lignin in sulphite wood pulp. Papierfabrik 8:1285–1286, Estimation of lignin in jute by titration method 5:521–522
Kong LY, Guo DS, Zhao BG, Li RG (2010) Preliminary purification and characterization of extracellular lignin peroxidase from Pseudomonas fluorescens GcM5-1A. Int J Autom Comput 32:112–116
Koschorreck K, Richter SM (2008) Cloning and characterization of a new laccase from Bacillus licheniformis catalyzing dimerization of phenolic acids. Appl Microbiol Biotechnol 79:217–224
Kratzl K, Vierhapper FW (1971) Spezifisch 14C-kernmarkierte Phenolderivate. 1. Mitt.: Synthese von 14C-Guajacol Mh. Chemistry 102:224–232
Kudo T (2009) Termite-microbe symbiotic system and its efficient degradation of lignocellulose. Biosci Biotechnol Biochem 73:2561–2567
Kuhad RC, Singh A, Eriksson KEL (1997) Microorganisms and enzymes involved in the degradation of plant fiber cell walls. Adv Biochem Eng Biotechnol 57:47–111
Kuhnigk T, König H (1997) Degradation of dimeric lignin model compounds by aerobic bacteria isolated from the hindgut of xylophagous termites. J Basic Microbiol 37:205–211
Kumar L, Rathore V, Srivastava H (2001) 14C-[lignin]-lignocellulose biodegradation by bacteria isolated from polluted soil. Indian J Exp Biol 39:584–589
Kuritz T, Wolk CP (1995) Use of filamentous Cyanobacteria for biodegradation of organic pollutants. Appl Environ Microbiol 61:234–238
Le Roes-Hill M, Khan N, Burton SG (2011) Actinobacterial peroxidases: an unexplored resource for biocatalysis. Appl Biochem Biotechnol 164:681–713
Lee B, Pometto AL, Fratzke A, Bailey TB (1991) Biodegradation of degradable plastic polyethylene by Phanerochaete and Streptomyces species. Appl Environ Microbiol 57:678–685
Leisola M, Pastinen O, Axe DD (2012) Lignin—designed randomness BIO-Complexity 2012 (3):1–11
Li RW, Connor EE, Li C, Baldwin RL, Sparks ME (2012) Characterization of the rumen microbiota of pre-ruminant calves using metagenomic tools. Environ Microbiol 14:129–139
Lilburn TG, Schmidt TM, Breznak JA (1999) Phylogenetic diversity of termite gut Spirochaetes. Environ Microbiol 1:331–345
Lim YW, Baik KS, Han SK, Kim SB, Bae KS (2003) Burkholderia sordidicola sp. nov., isolated from the white-rot fungus Phanerochaete sordida. Int J Syst Evol Microbiol 53:1631–1636
Lu W-J, Wang H-T, Nie Y-F, Wang Z-C, Huang D-Y, Qiu X-Y, Chen J-C (2004) Effect of inoculating flower stalks and vegetable waste with ligno-cellulolytic microorganisms on the composting process. J Environ Sci Health B 39:871–887
Lü F, Bize A, Guillot A, Monnet V, Madigou C, Chapleur O, Mazéas L, He P, Bouchez T (2013) Metaproteomics of cellulose methanisation under thermophilic conditions reveals a surprisingly high proteolytic activity. ISME J 8:88–102
Maeda M, Chung S-Y, Song E, Kudo T (1995) Multiple genes encoding 2, 3-dihydroxybiphenyl 1, 2-dioxygenase in the gram-positive polychlorinated biphenyl-degrading bacterium Rhodococcus erythropolis TA421, isolated from a termite ecosystem. Appl Environ Microbiol 61:549–555
Magliozzo RS, Marcinkeviciene JA (1997) The role of Mn(II)-peroxidase activity of mycobacterial catalase-peroxidase in activation of the antibiotic isoniazid. J Biol Chem 272:8867–8870
Malachowsky K, Phelps T, Teboli A, Minnikin D, White D (1994) Aerobic mineralization of trichloroethylene, vinyl chloride, and aromatic compounds by Rhodococcus species. Appl Environ Microbiol 60:542–548
Manter DK, Hunter WJ, Vivanco JM (2011) Enterobacter soli sp. nov.: a lignin-degrading γ-Proteobacteria isolated from soil. Curr Microbiol 62:1044–1049
Martínková L, Uhnáková B, Pátek M, Nešvera J, Křen V (2009) Biodegradation potential of the genus Rhodococcus. Environ Int 35:162–177
Martins LO, Soares CM (2002) Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J Biol Chem 277:18849–18859
Marton J, Marton T (1964) Molecular weight of kraft lignin. TAPPI J 47:471–476
Masai E, Shinohara S, Hara H, Nishikawa S, Katayama Y, Fukuda M (1999) Genetic and biochemical characterization of a 2-pyrone-4, 6-dicarboxylic acid hydrolase involved in the protocatechuate 4, 5-cleavage pathway of Sphingomonas paucimobilis SYK-6 J. Bacteriology 181:55–62
Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem 71:1–15
Mathew GM, Ju YM (2011) Microbial community analysis in the termite gut and fungus comb of Odontotermes formosanus: the implication of Bacillus as mutualists. FEMS Microbiol Ecol 79:504–517
Mathieu Y, Gelhaye E, Dumarcay S, Gerardin P, Harvengt L, Buee M (2013) Selection and validation of enzymatic activities as functional markers in wood biotechnology and fungal ecology. J Microbiol Methods 92:157–163
McCarthy AJ (1987) Lignocellulose-degrading actinomycetes. FEMS Microbiol Rev 46:145–163
Meentemeyer V (1978) Macroclimate and lignin control of litter decomposition rates. Ecology 59:465–472
Meux E, Prosper P, Masai E, Mulliert G, Dumarçay S, Morel M, Didierjean C, Gelhaye E, Favier F (2012) Sphingobium sp. SYK-6 LigG involved in lignin degradation is structurally and biochemically related to the glutathione transferase omega class. FEBS Lett 586:3944–3950
Mikesková H, Novotný Č, Svobodová K (2012) Interspecific interactions in mixed microbial cultures in a biodegradation perspective. Appl Microbiol Biotechnol 95:861–870
Mitsui R, Kusano Y, Kurimoto H, Sakai Y, Kato N, Tanaka M (2003) Formaldehyde fixation contributes to detoxification for growth of a nonmethylotroph, Burkholderia cepacia TM1, on vanillic acid. Appl Environ Microbiol 69:6128–6132
Morozova OV, Shumakovich GP, Shleev SV, Yaropolov YI (2007) Laccase-mediator systems and their applications: a review. Appl Biochem Microbiol 43:523–535
Moya R, Hernandez M (2009) Contributions to a better comprehension of redox-mediated decolouration and detoxification of azo dyes by a laccase produced by Streptomyces cyaneus CECT 3335. Bioresour Technol 101:2224–2229
Müller-Enoch D, Thomas H, Holzmann P, Haider K, Haider H (1974) Metabolisierung von 3,4-Dimethoxybenzaldehydund 3,4-Dimethoxybenzoesäiure in der isoliert perfundierten Rattenleber. Z Naturforsch 29c:602–607
Nakata K (2000) High resistance to oxygen radicals and heat is caused by a galactoglycerolipid in Microbacterium sp. M874. J Biochem 127:731–737
Ni JF, Tokuda G (2013) Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol Adv 31:838–850
Niladevi KN, Prema P (2005) Mangrove actinomycetes as the source of ligninolytic enzymes. Actinomycetologica 19:40–47
Nimchua T, Uengwetwanit T, Eurwilaichitr L (2012) Metagenomic analysis of novel lignocellulose-degrading enzymes from higher termite guts inhabiting microbes. J Microbiol Biotechnol 22:462–469
Nishikawa NK, Sutcliffe R, Saddler JN (1988) The influence of lignin degradation products on xylose fermentation by Klebsiella pneumoniae. Appl Microbiol Biotechnol 27:549–552
Odier E, Monties B (1977) Activite ligninolytique in vitro de bacteries isolees de paille de Ble en decomposition. C R Acad Sci D 284:2175–2178
Odier E, Monties B (1978) Biodegradation de la lignine de blé par Xanthomonas 23. Annales de l'Institut Pasteur/Microbiologie 129A:361–377
Okino LK, Machado KMG, Fabris C, Bononi VLR (2000) Ligninolytic activity of tropical rainforest basidiomycetes. World J Microbiol Biotechnol 16:889–893
Oliveira PL, Duarte MCT, Ponezi AN, Durrant LR (2009) Purification and Partial characterization of manganese peroxidase from Bacillus pumilus and Paenibacillus sp. Braz J Microbiol 40:818–826
Pagani I, Liolios K, Jansson J, Chen I-MA, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC (2012) The genomes online database (GOLD) v. 4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 40:D571–D579
Pason P, Kyu KL, Ratanakhanokchai K (2006) Paenibacillus curdlanolyticus strain B-6 xylanolytic-cellulolytic enzyme system that degrades insoluble polysaccharides. Appl Environ Microbiol 72:2483–2490
Patrauchan MA, Florizone C, Eapen S, Gómez-Gil L, Sethuraman B, Fukuda M, Davies J, Mohn WW, Eltis LD (2008) Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1. J Bacteriol 190:37–47
Paulsen IT, Seshadri R, Nelson KE, Eisen JA, Heidelberg JF, Read TD, Dodson RJ, Umayam L, Brinkac LM, Beanan MJ (2002) The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc Natl Acad Sci 99:13148–13153
Pellinen J, Vfiisfinen E, Salkinoja-Salonen M, Brunow G (1984) Utilization of dimeric lignin model compounds by mixed bacterial cultures. Appl Microbiol Biotechnol 20:77–82
Peng X, Egashira T, Hanashiro K, Masai E, Nishikawa S, Katayama Y, Kimbara K, Fukuda M (1998) Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl Environ Microbiol 64:2520–2527
Perestelo F, Falcon MA (1989) Bioalteration of kraft pine lignin by Bacillus megaterium isolated from compost piles. J Ferment Bioeng 68:151–153
Perestelo F, Rodriguez A, Pérez R, Carnicero A, de la Fuente G, Falcon MA (1996) Short communication: isolation of a bacterium capable of limited degradation of industrial and labelled, natural and synthetic lignins. World J Microbiol Biotechnol 12:111–112
Raghukumar C, Vipparty V, David JJ, Chandramohan D (2001) Degradation of crude oil by marine cyanobacteria. Appl Microbiol Biotechnol 57:433–436
Raj A, Reddy MK, Chandra R (2007) Decolourisation and treatment of pulp and paper mill effluent by lignin-degrading Bacillus sp. J Chem Technol Biotechnol 82:399–406
Rencoret J, Marques G, Guiérrez A, Lidia N, Jiménez-Barbero J, Martinez AT, Rio JC (2009) Isolation and structural characterization of the milled-wood lignin from Paulownia fortunei wood. Ind Crop Prod 30:137–143
Ruijssenaars HJ, Hartmans S (2004) A cloned Bacillus halodurans multicopper oxidase exhibiting alkaline laccase activity. Appl Microbiol Biotechnol 65:177–182
Sakai M, Ezaki S, Suzuki N, Kurane R (2005) Isolation and characterization of a novel polychlorinated biphenyl-degrading bacterium, Paenibacillus sp. KBC101. Appl Microbiol Biotechnol 68:111–116
Santos A, Mendes S, Brissos V, Martins LO (2013) New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: towards biotechnological applications. Appl Microbiol Biotechnol 98(5):2053–2065
Sarkanen KV, Ludwig CH (1971) Lignins: occurrence, formation, structure and reactions. Wiley-Interscience, New York, pp 673–675
Satpathy R, Behera R, Padhi S, Guru R (2013) Computational phylogenetic study and data mining approach to laccase enzyme sequences. J Phylogenet Evol Biol 1:2
Schneider T, Keiblinger KM, Schmid E, Sterflinger-Gleixner K, Ellersdorfer G, Roschitzki B, Richter A, Eberl L, Zechmeister-Boltenstern S, Riedel K (2012) Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions. ISME J 6:1749–1762
Seto M, Kimbara K, Shimura M, Hatta T, Fukuda M, Yano K (1995) A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl Environ Microbiol 61:3353–3358
Sharma P, Goel R, Capalash N (2007) Bacterial laccases. World J Microbiol Biotechnol 23:823–832
Shashirekha S, Uma L, Subramanian G (1997) Phenol degradation by the marine cyanobacterium Phormidium valderianum BDU 30501. J Ind Microbiol Biotechnol 19:130–133
Shi Y, Chai L, Tang C, Yang Z, Zheng Y, Chen Y, Jing Q (2013) Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioprocess Biosyst Eng 36(12):1957–1965
Song YJ (2009) Characterization of aromatic hydrocarbon degrading bacteria isolated from pine litter. Korean J Microbiol Biotechnol 37:333–339
Sutherland JB, Blanchette RA, Crawford DL, Pometto AL (1979) Breakdown of Douglas fir phoem by a lignocellulose degrading Streptomyces. Curr Microbiol 2:123–126
Tan K, Chang C, Cuff M, Osipiuk J, Landorf E, Mack JC, Zerbs S, Joachimiak A, Collart FR (2013) Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-coumaric acid and related aromatic acids. Proteins Struct Funct Bioinforma 81:1709–1726
Taylor CR, Hardiman EM, Ahmad M, Sainsbury PD, Norris PR, Bugg TDH (2012) Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J Appl Microbiol 113:521–530
Treadway SL, Yanagimachi KS, Lankenau E, Lessard PA, Sinskey AJ (1999) Isolation and characterization of indene bioconversion genes from Rhodococcus strain I24. Appl Microbiol Biotechnol 51:786–793
Tsang A, Butler G, Powlowski J, Panisko EA, Baker SE (2009) Analytical and computational approaches to define the Aspergillus niger secretome. Fungal Genet Biol 46:S153–S160
Uthandi S, Saad B, Humbard MA, Maupin-Furlow JA (2010) LccA, an Archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax volcanii. Appl Environ Microbiol 76(3):733–743
van der Lelie D, Taghavi S, McCorkle SM, Li L-L, Malfatti SA, Monteleone D, Donohoe BS, Ding S-Y, Adney WS, Himmel ME (2012) The metagenome of an anaerobic microbial community decomposing poplar wood chips. PLoS One 7:e36740
Vasin A, Klotchenko S, Puchkova L (2013) Phylogenetic analysis of six-domain multi-copper blue proteins. Plos Curr Tree Life
Viñas M, Sabaté J, Espuny MJ, Solanas AM (2005) Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol 71:7008–7018
Wang Y, Liu Q, Yan L, Gao Y, Wang Y, Wang W (2013) A novel lignin degradation bacterial consortium for efficient pulping. Bioresour Technol 139:113–119
Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565
Wegener G, Fengel D (1977) Studies on milled wood lignins from spruce part 1. Composition and molecular properties. Wood Sci Technol 11:133–145
Williams PA, Murray K (1974) Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol 120:416–423
Wischgoll S, Heintz D, Peters F, Erxleben A, Sarnighausen E, Reski R, Dorsselaer AV, Boll M (2005) Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol Microbiol 58:1238–1252
Wittich RM, Wilkes H, Sinnwell V, Francke W, Fortnagel P (1992) Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl Environ Microbiol 58:1005–1010
Wu YR, He J (2013) Characterization of anaerobic consortia coupled lignin depolymerization with biomethane generation. Bioresour Technol 139:5–12
Zeng J, Lin X, Zhang J, Li X, Wong MH (2011) Oxidation of polycyclic aromatic hydrocarbons by the bacterial laccase CueO from E. coli. Appl Microbiol Biotechnol 89(6):1841–1849
Zerbini JE, Oliveira MM, Bon EPS (1999) Lignin peroxidase production by Streptomyces viridosporus T7A, vol 77–79. Humana Press Inc., Rio de Janeiro
Zhong WZ, Zhang ZZ, Luo YJ, Sun SS, Qiao W, Xiao M (2011) Effect of biological pretreatments in enhancing corn straw biogas production. Bioresour Technol 102:11177–11182
Zhou JT, Guan XY, Qu YY, Li A, Gou M, Ai FF (2010) Research on isolation, identification of a phenol-degrading strain Brucella sp. GXY-1 and characteristics of its degradation and crude enzyme. Dalian Ligong Daxue Xuebao/J Dalian Univ Technol 50:340–345
Acknowledgments
This work was supported by a grant from the “French Environment and Energy Agency, ADEME” (Project No. 13 06 C 0068).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tian, JH., Pourcher, AM., Bouchez, T. et al. Occurrence of lignin degradation genotypes and phenotypes among prokaryotes. Appl Microbiol Biotechnol 98, 9527–9544 (2014). https://doi.org/10.1007/s00253-014-6142-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6142-4